We are a user-driven database of open neuroscience projects
What we do

Project curators
We curate projects from the web and allow creators to register their own

Extensive database
We make it easy for researchers to find relevant projects to help their studies

Project promoters
We help creators and promote their work through our network
Projects
Want to add your own? Click below!

A place to discuss experimental methods in animal behaviour
TheBehaviourForum.org (www.thebehaviourforum.com) is a global platform bringing together scientists, technicians, and animal welfare experts to advance the study of animal behaviour. This free, user-friendly space fosters collaboration, enabling members to share knowledge, troubleshoot experiments, and discuss best practices.
Bombcell: automated quality control for extracellular electrophysiology
Recent advances in high-density neural probes, such as Neuropixels, have revolutionized electrophysiology. However, they produce large, complex datasets that require extensive quality control. This is typically done through manual curation, which is time-consuming, subjective, and lacks standardization.

Democratizing Smart Microscopy with navigate
Neuroscientific imaging demands advanced techniques capable of resolving intricate neural circuits, fine axonal structures, and precise synaptic connections within expansive, three-dimensional volumes. Light-sheet fluorescence microscopy has emerged as a transformative tool for this purpose, enabling high-speed, three-dimensional imaging with minimal photodamage.
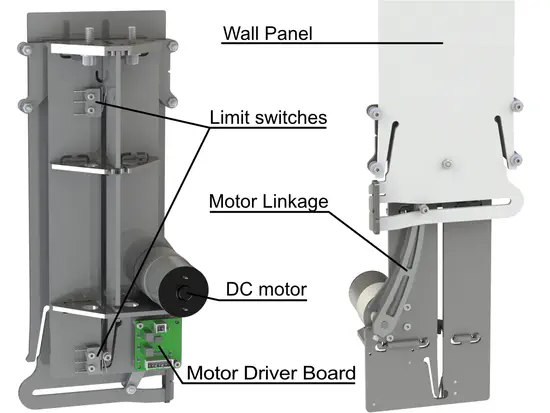
NC4Gate: A modular gate system for autonomous control of rodent behavior
Rodent mazes have been used for decades to study the neural basis of behavior. Advancements in rapid prototyping techniques and access to affordable electronics allows laboratories with sufficient expertise in engineering and programming to customize and construct maze apparatuses and behavioral tasks, thereby increasing the ability of their studies to answer specific scientific questions.

Neurobagel
Neurobagel aims to connect decentralized neuroimaging datasets using linked data principles while allowing data files and access control to remain with research institutes. The Neurobagel software ecosystem provides reusable data schemas and intuitive tools to:

NeuroML
NeuroML is an international, collaborative initiative to develop a language and associated software ecosystem for working with data-driven biophysically detailed models of neural systems. With NeuroML, you can easily create, visualise, analyse, inspect, simulate, share, and re-use models and model components.

Towards open meta-research in neuroimaging
When meta-research (research on research) makes an observation or points out a problem (such as a flaw in methodology), the project should be repeated later to determine whether the problem remains.
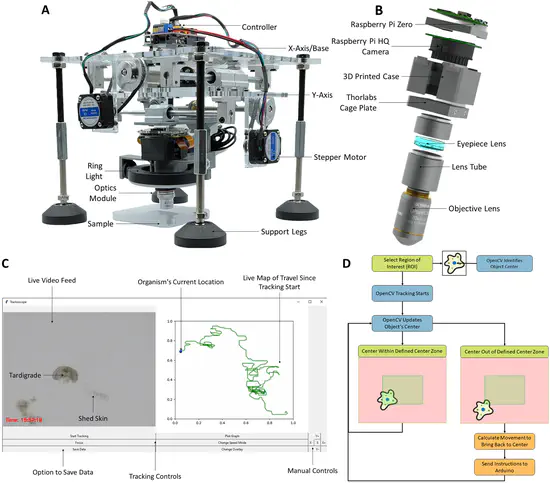
Trackoscope
World Wide Series Seminar Cells and microorganisms are motile, yet the stationary nature of conventional microscopes impedes comprehensive, long-term behavioral and biomechanical analysis. The limitations are twofold: a narrow focus permits high-resolution imaging but sacrifices the broader context of organism behavior, while a wider focus compromises microscopic detail.

BORIS
BORIS is an easy-to-use event logging software for video/audio coding and live observations. Project Author(s) Olivier Friard; Marco Gamba Project Links www.boris.unito.it This post was automatically generated by anonymous

TWINKLE: An open-source two-photon microscope for teaching and research
We present “Twinkle”: a microscope for Two-photon Imaging in Neuroscience, and Kit for Learning and Education. It is a fully open, high-performance and cost-effective research and teaching microscope without any custom parts beyond what can be fabricated in a university machine shop.
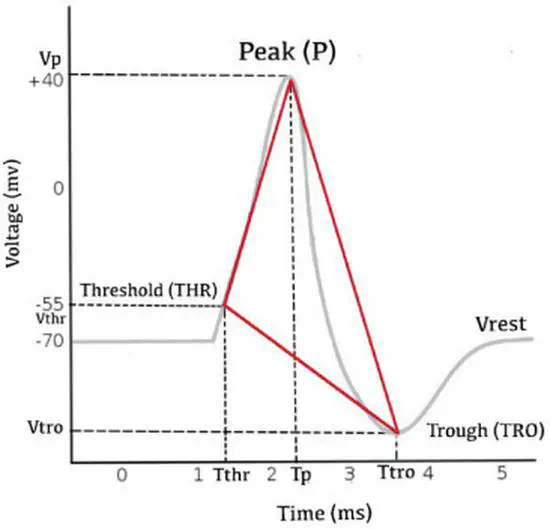
Neuronal Spike Shapes (NSS), a simple approach for analyzing the electrophysiological profiles of cells based on their Action Potential (AP) waveforms.
This repository contains the code of the Neuronal Spike Shapes (NSS), a simple approach for analyzing the electrophysiological profiles of cells based on their Action Potential (AP) waveforms. The NSS method explores the heterogeneity of cell types and states by summarizing the AP waveform into a triangular representation complemented by a set of derived electrophysiological (EP) features.

NeuroAnsible
NeuroAnsible is a software installation system to take an Ubuntu LTS desktop and turn it into a Neuroimaging workstation. NeuroAnsible installs software in a module system similar to those available on large HPCs, aiming to be minimally invasive to the underlying system.
BCIWiki
BCIWiki is a collection of links and resources related to computers that interface with the nervous system. The long-term goals for this project are to make it simple for anyone to understand the capabilities and limitations of Brain-Computer Interface (BCI) technologies, and to make informed hypotheses about their potential uses.
NeuroMatic
NeuroMatic is a collection of WaveMetrics Igor Pro software tools for acquiring, analyzing and simulating electrophysiological data. By merging a wide range of tools into a single package, NeuroMatic facilitates a more integrated style of research.
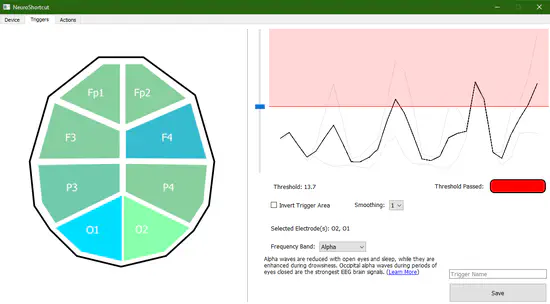
NeuroShortcut
NeuroShortcut is a brain-computer interface tool for triggering keyboard shortcuts, system commands, and macro scripts by changing mental states. The application uses the Brainflow library to stream live band power data to an interface where a user can then save a threshold value for triggering events on their computer.
Chronic Recoverable Neuropixels in Mice
This protocol collection explains how to build a low-cost, lightweight system to implant 1 Neuropixels 1.0 probe or 2 Neuropixels 2.0 probes into mice, record during freely moving behavior, then recover the probes for future use.

iElectrodes
iElectrodes is an open-source Matlab toolbox to localize intracranial electrodes from MRI and CT images. Atlas and brain surfaces can be easily loaded for anatomical labeling. The toolbox is capable of working with ECoG (subdural grids and strips) and SEEG (depth) electrodes.

DataJoint Elements
DataJoint Elements is a growing compilation of community-curated, open-source software modules for building automated data pipelines and analysis workflows for neuroscience experiments. DataJoint Elements enables: Secure tracking of animal subjects, equipment, and procedures Automatic data ingestion Customized research workflows Faster deployment Improved reproducibility Greater continuity across projects Secure tracking.

MagnetSearch
A search for magnetosensitive neurons Many species across the animal kingdom are said to use the Earth’s magnetic field for orientation and navigation. However, the mechanisms by which information about magnetic fields enters the nervous system remain unknown.

Pi USB Cam - An Affordable, Simple, Powerful, & Versatle DIY Video Recording Solution for Behavioral Neuroscientists
Video capture is increasingly necessary for neuroscience research where neural and behavioral data are synchronized to reveal correlative and causal relationships. This relies on a recording system that can capture quality videos without significant alterations to preexisting experimental conditions (e.

DataJoint Core
DataJoint Core is an open-source toolkit for defining and operating computational data pipelines (i.e., sequences of steps for data acquisition, processing, and transformation). Pipelines built in DataJoint Core offer: Efficient design with intuitive queries Automated, reproducible computation with full referential integrity Coordination of multiple human and computer workers Flexibility to adapt and change DataJoint Core includes libraries for Python and MATLAB, a REST API, and GUI tools for data entry and visualizations.

SqueakR: An Experiment Interface for ‘DeepSqueak’ Bioacoustics Research
SqueakR is an open-source R package, available on CRAN, which streamlines bioacoustics research through automated data processing and visualizations for rodent vocalizations exported from DeepSqueak. These functions are harnessed through the ‘SqueakR’ Shiny Dashboard, available as a function within the package, which can be used to visualize experimental results and analyses, as well as conduct statistical significance test between call features across groups.
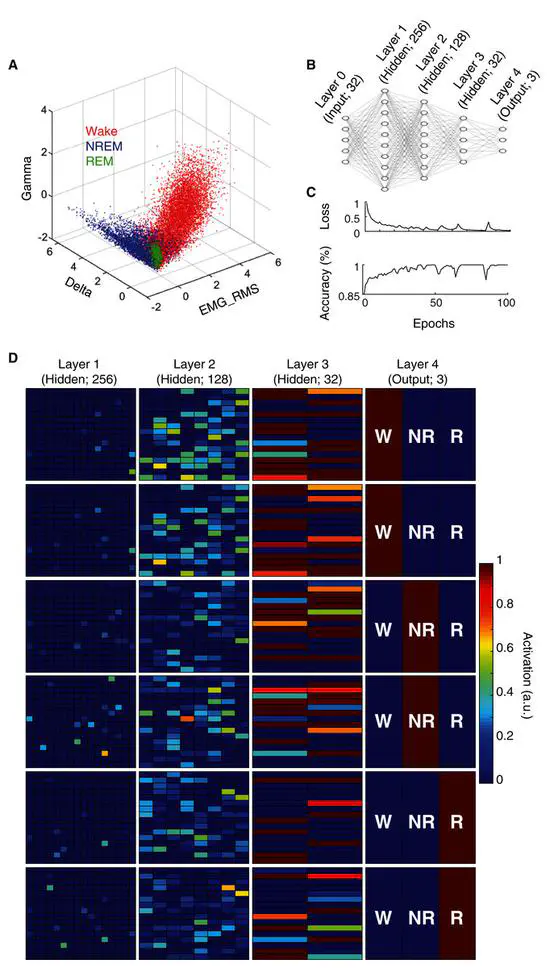
An artificial neural network for automated behavioral state classification in rats
While accurate behavioral state classification is critical for many research applications, it is often done manually, which can be both tedious and inaccurate. Here we present a novel artificial neural network that uses electrophysiological features to automatically classify behavioral state in rats with high accuracy, sensitivity, and specificity.
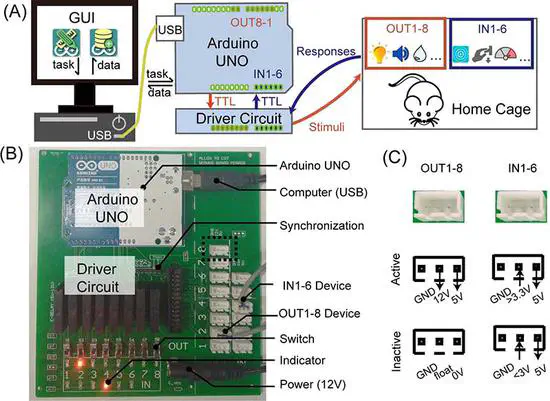
ArControl - Arduino Digital I/O System
ArControl is a Arduino based digital signals control system. A special application for ArControl is to establish a animal behavioral platform (as Skinner box), which control devices to deliver stimulation, monitor behavioral response and record data.
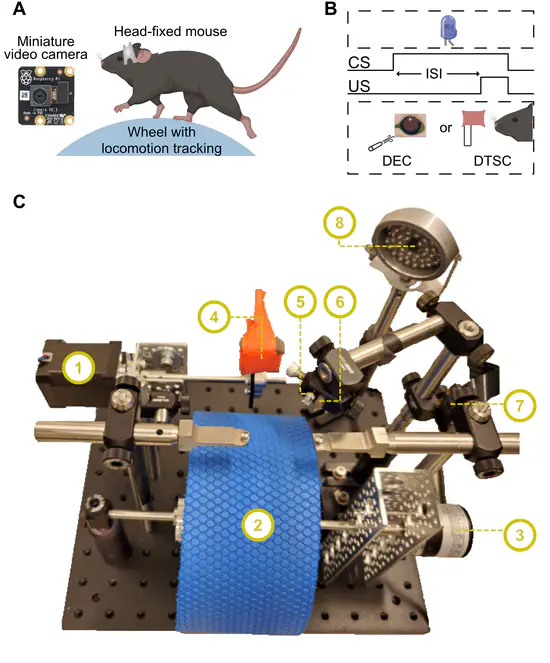
A Flexible Platform for Monitoring Cerebellum-Dependent Sensory Associative Learning
Climbing fiber inputs to Purkinje cells provide instructive signals critical for cerebellum-dependent associative learning. Studying these signals in head-fixed mice facilitates the use of imaging, electrophysiological, and optogenetic methods. Here, a low cost behavioral platform (~$1000) was developed that allows tracking of associative learning in head-fixed mice that locomote freely on a running wheel.
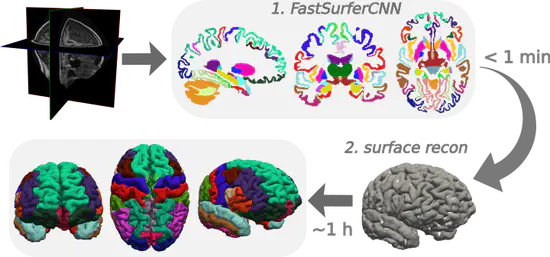
FastSurfer
FastSurfer is a fast and extensively validated deep-learning pipeline for the fully automated processing of structural human brain MRIs. As such, it provides FreeSurfer conform outputs, enables efficient big-data analysis for large cohort studies, and time-critical clinical applications such as structure localization during image acquisition or rapid extraction of quantitative measures for precision medicine.
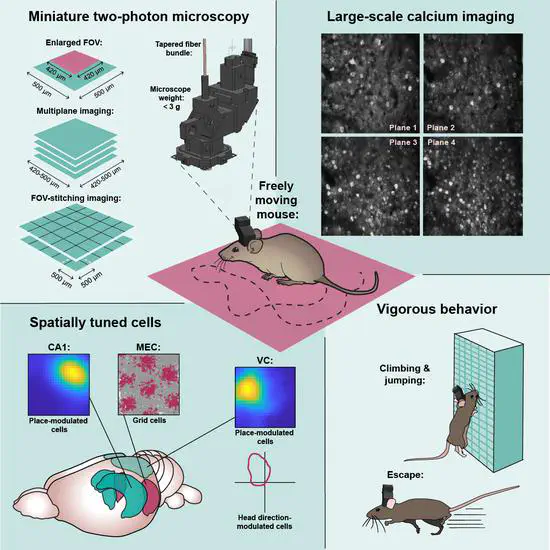
MINI2P
MINI2P is an open-source miniature 2-photon microscope brain explorer for fast high-resolution calcium imaging in freely-moving mice. In brief Development of a miniature 2-photon miniscope for large-scale calcium imaging in freely moving mice allows stable simultaneous recording of more than a thousand cells across multiple planes of densely active cortical regions in a wide spectrum of behavioral tasks without impediment of the animal’s behavior.

Optogenetics TTL driver
This simple optogenetics stimulation driver is based on the Arduino Uno, and produces millisecond-accurate timing to control a single laser or LED. The dials allow easy control of pulse on-time and frequency, with a simple switch to turn the flashing on or off.
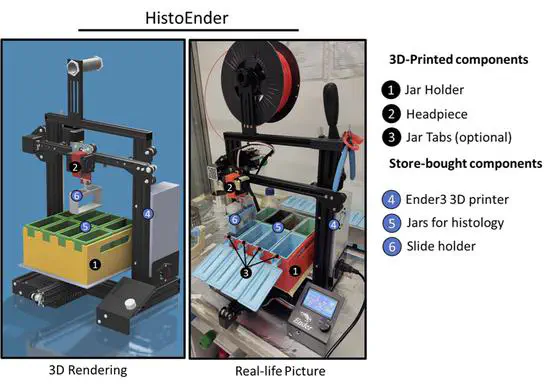
HistoEnder
Automated microscope slide stainers are typically very expensive, and unless the lab performs heavy histological work it is hard to justify buying a €2000-€10000 machine. As a result, histology and pathology labs around the world lose thousands of working hours for following a procedure that could be easily automated.
Mesmerize
Mesmerize is a platform for the annotation and analysis of neuronal calcium imaging data. Mesmerize encompasses the entire process of calcium imaging analysis from raw data to interactive visualizations. Mesmerize allows you to create FAIR-functionally linked datasets that are easy to share.
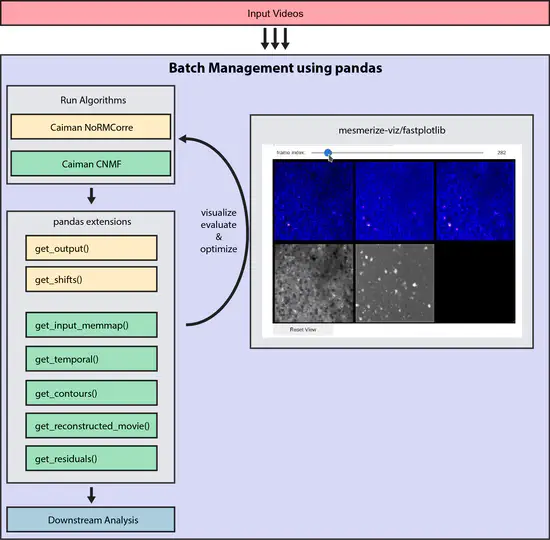
Mesmerize Core
A batch management system for calcium imaging analysis using the CaImAn library. It contains pandas.DataFrame and pandas.Series extensions that interface with CaImAn for running the various algorithms and organizing input & output data.
Raspberry Pi controlled precise treat dispenser
Some forms of canine cognition research require a dispenser that can accurately dispense precise quantities of treats. When using off-the-shelf or retrofitted dispensers, there is no guarantee that a precise number of treats will be dispensed.
TopMouseTracker
TopMouseTacker is a Python library based on OpenCV made for object tracking in videos. It uses a color camera and depth camera to infer the position of objects in the arena.

CaImAn: open source scalable algorithms for calcium and voltage imaging data
Advances in fluorescence microscopy enable monitoring larger brain areas in-vivo with finer time resolution. The resulting data rates require reproducible analysis pipelines that are reliable, fully automated, and scalable to datasets generated over the course of months.

COGAIN Association (COmmunication by GAze INteraction)
The COGAIN Association aims to promote research and development in the field of gaze-based interaction in computer-aided communication and control. Computer applications can be controlled by gazing at the computer screen.
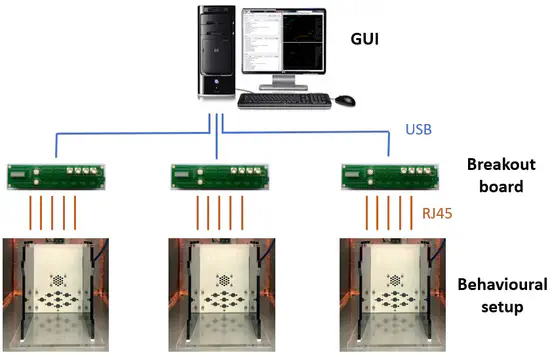
PyControl
Open source, Python based, behavioural experiment control. pyControl is a system of open source hardware and software for controlling behavioural experiments, built around the Micropython microcontroller. pyControl makes it easy to program complex behavioural tasks using a clean, intuitive, and flexible syntax for specifying tasks as state machines.
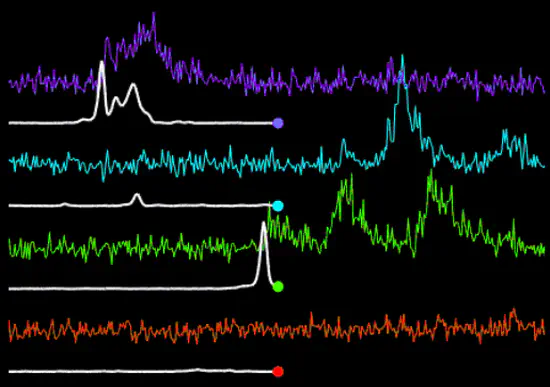
CASCADE: Calibrated inference of spiking from calcium ΔF/F data using deep networks
CASCADE translates calcium imaging ΔF/F traces into spiking probabilities or discrete spikes. CASCADE is based on deep networks and runs on Python (Linux/Windows/Mac); it can be used either as a web application (Colaboratory notebook), or locally on a CPU or GPU.

DANNCE
DANNCE (3-Dimensional Aligned Neural Network for Computational Ethology) is a convolutional neural network (CNN) that calculates the 3D positions of user-defined anatomical landmarks on behaving animals from videos taken at multiple angles.
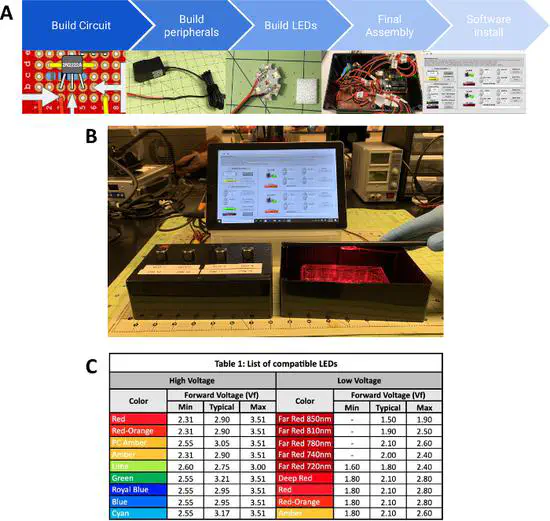
Building a Simple and Versatile Illumination System for Optogenetic Experiments
Controlling biological processes using light has increased the accuracy and speed with which researchers can manipulate many biological processes. Optical control allows for an unprecedented ability to dissect function and holds the potential for enabling novel genetic therapies.

GeNN
GeNN is a GPU enhanced Neuronal Network simulation environment supporting multiple frontends and backends. Spiking neural network models can be specified directly in Python or C++ or through interfaces to Brian 2, SpineCreator (SpineML), or PyNN.

Pingouin
Pingouin is an open-source statistical package written in Python 3 and based mostly on Pandas and NumPy. Some of its main features are listed below. For a full list of available functions, please refer to the API documentation.

YASA
YASA (Yet Another Spindle Algorithm) is a command-line sleep analysis toolbox in Python. The main functions of YASA are: Automatic sleep staging of polysomnography data (see preprint article). Event detection: sleep spindles, slow-waves and rapid eye movements, on single or multi-channel EEG data.

DANDI: A data archive and collaboration space for neurophysiology
The US BRAIN Initiative archive for publishing and sharing neurophysiology data including electrophysiology, optophysiology, and behavioral time-series, and images from immunostaining experiments. DANDI: Distributed Archives for Neurophysiology Data Integration is a Web platform for scientists to share, collaborate, and process data from cellular neurophysiology experiments.

GuPPy, a Python toolbox for the analysis of fiber photometry data
Fiber photometry (FP) is an adaptable method for recording in vivo neural activity in freely behaving animals. It has become a popular tool in neuroscience due to its ease of use, low cost, the ability to combine FP with freely moving behavior, among other advantages.
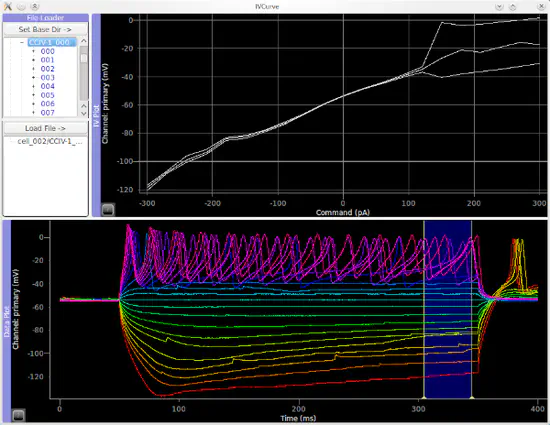
ACQ4
ACQ4 is a python-based platform for experimental neurophysiology developed at the Allen Institute for Brain Science and the University of North Carolina, Chapel Hill. It includes support for patch-clamp electrophysiology, multiphoton imaging, scanning laser photostimulation, and many other experimental techniques.
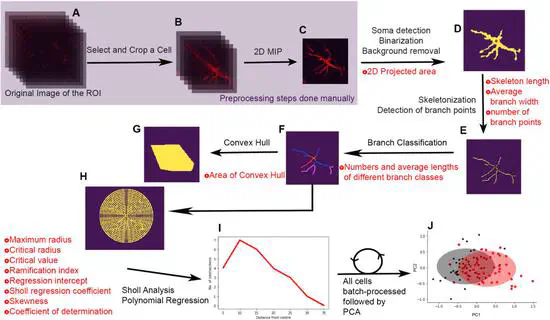
Automated morphometric analysis with SMorph software reveals plasticity induced by antidepressant therapy in hippocampal astrocytes
Nervous system development and plasticity involves changes in cellular morphology, making morphological analysis a valuable exercise in the study of nervous system development, function and disease. Morphological analysis is a time-consuming exercise requiring meticulous manual tracing of cellular contours and extensions.
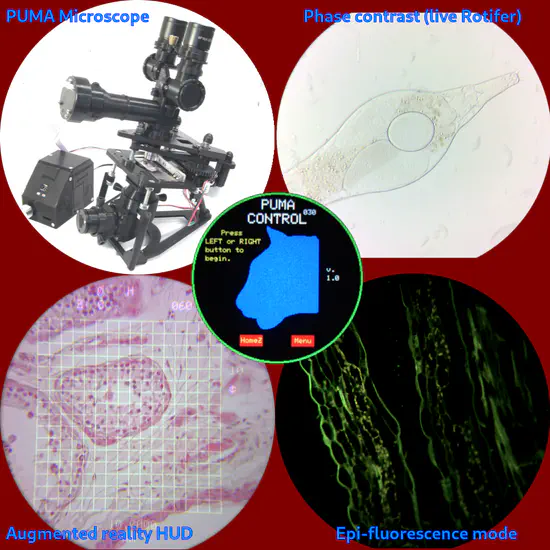
PUMA 3D Printed Open Source Microscope
PUMA is 3D printed open source portable modular microscope with augmented reality, fluorescence, phase contrast, epi-illumination, epi- and trans-polarisation and other features. You can either build it fully DIY or purchase the basic scope ready made and upgrade it as required (see below for details).

The Open-Source UCLA Miniscope Project
One of the biggest challenges in neuroscience is to understand how neural circuits in the brain process, encode, store, and retrieve information. Meeting this challenge requires tools capable of recording and manipulating the activity of intact neural networks in freely behaving animals.
Diffusion MRI Q-ball CSA-ODF and Hough Tractography
This is a diffusion-weighted MRI processing Matlab toolbox (including binaries), which can be used to: • Compute the Q-Ball Imaging Orientation Distribution Function in Constant Solid Angle (CSA-ODF) (Aganj et al, MRM 2010).
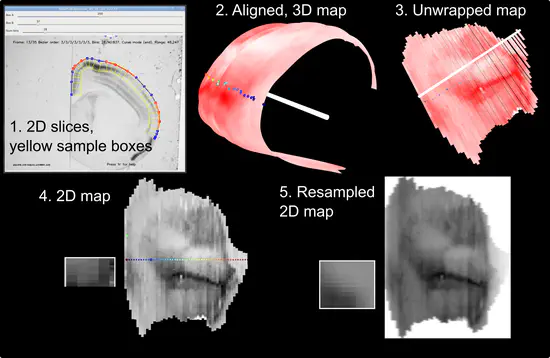
Stalefish
Stalefish is a tool to analyse gene expression from ISH or other histology slice images. From your 2D brain slice images you can build a 3D expression surface from a brain structure such as the isocortex or the hippocampus.

Autopilot
Autopilot is a Python framework for performing complex, hardware-intensive behavioral experiments with swarms of networked Raspberry Pis. As a tool, it provides researchers with a toolkit of flexible modules to design experiments without rigid programming & API limitations.

BioAmp EXG Pill
BioAmp EXG Pill is a small, powerful Analog Front End (AFE) biopotential signal acquisition board that can be paired with any 5 V Micro Controller Unit (MCU) with an ADC. It is capable of recording publication-quality biopotential signals like ECG, EMG, EOG, and EEG, without the inclusion of any dedicated hardware or software filters.

NeuroDesk
Neuroimaging researchers require a diverse collection of bespoke command-line and graphical tools to analyse data and answer research questions. Installing and maintaining a neuroimaging software setup is challenging and often results in un-reproducible environments.
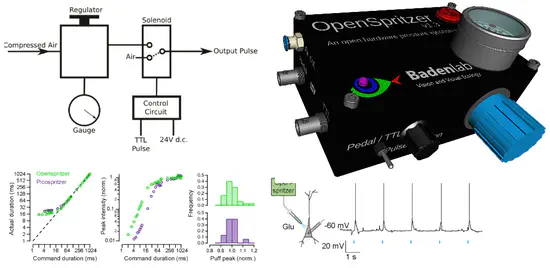
OpenSpritzer
Designed for ease of use, robustness and low-cost, the “Openspritzer” is an open hardware “Picospritzer” as routinely used in biological labs around the world. The performance of Openspritzer and commercial alternatives is effectively indistinguishable.
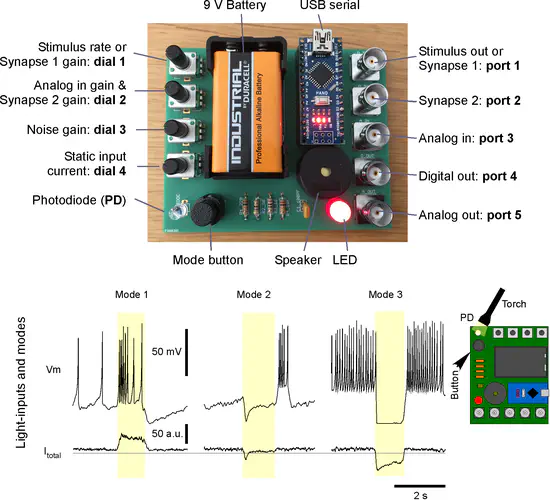
Spikeling
Understanding of how neurons encode and compute information is fundamental to our study of the brain, but opportunities for hands-on experience with neurophysiological techniques on live neurons are scarce in science education.

Clinica
Clinica is a software platform for clinical research studies involving patients with neurological and psychiatric diseases and the acquisition of multimodal data (neuroimaging, clinical and cognitive evaluations, genetics…), most often with longitudinal follow-up.
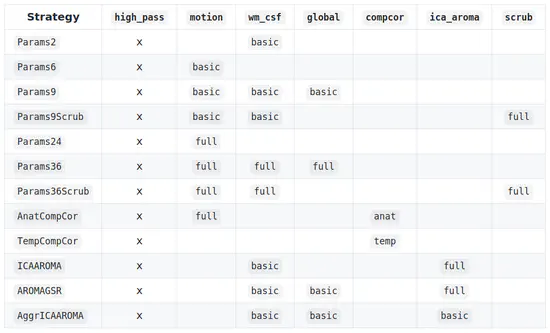
Load confound
Load a sensible subset of the fMRI confounds generated with fMRIprep in python (Esteban et al., 2018). The predefined denoising strategies are all adapted from Ciric et al. 2017. Check the docstring of each strategy for more info and a list of parameters.

Mobilefuge: Low-cost, Open-source 3D-printed centrifuge
We made a low-cost centrifuge that can be useful for carrying out low-cost LAMP based detection of SARS-Cov2 virus in saliva. The 3D printed centrifuge (Mobilefuge) is portable, robust, stable, safe, easy to build and operate.

The Virtual Macaque Brain
A whole-cortex macaque structural connectome constructed from a combination of axonal tract-tracing and diffusion-weighted imaging data. Created for modeling brain dynamics using TheVirtualBrain (thevirtualbrain.org) platform. A detailed description and example usage can be found in the paper here: https://www.

NEUROTWIN
The NEUROTWIN project is a scientific collaboration between the Transylvanian Institute of Neuroscience (TINS) from Cluj-Napoca, Romania, and three internationally acclaimed European institutions. The project is funded through the Twinning instrument, call H2020-WIDESPREAD-2020-5, of the European Union.
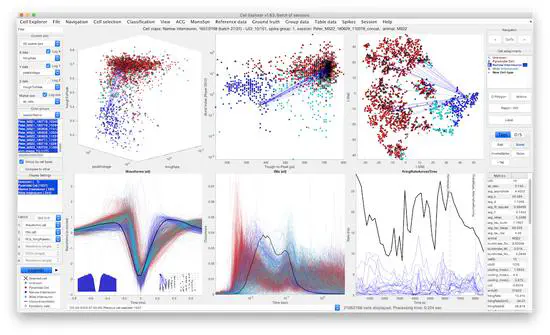
CellExplorer - Framework for analyzing single cells
The large diversity of cell-types of the brain, provides the means by which circuits perform complex operations. Understanding such diversity is one of the key challenges of modern neuroscience. Neurons have many unique electrophysiological and behavioral features from which parallel cell-type classification can be inferred.

BrainGlobe
World Wide Series Seminar BrainGlobe is a suite of Python-based computational neuroanatomy software tools. We provide software packages for the analysis and visualisation of neuroanatomical data, particularly from whole-brain microscopy. In addition, we provide tools for working with brain atlases, to simplify development of new tools and aid collaboration and cooperation by adopting common standards.
Kilosort
World Wide Series Seminar Kilosort is a software package for identifying neurons and their spikes in extracellular electrophysiology, a process known as “spike sorting”. Kilosort has been primarily developed and tested on the Neuropixels 1.
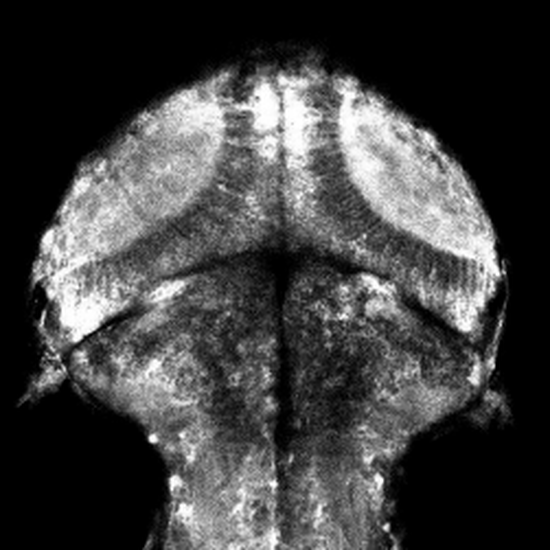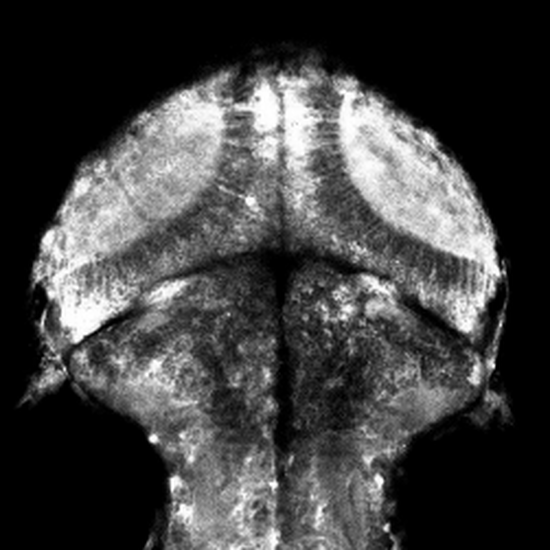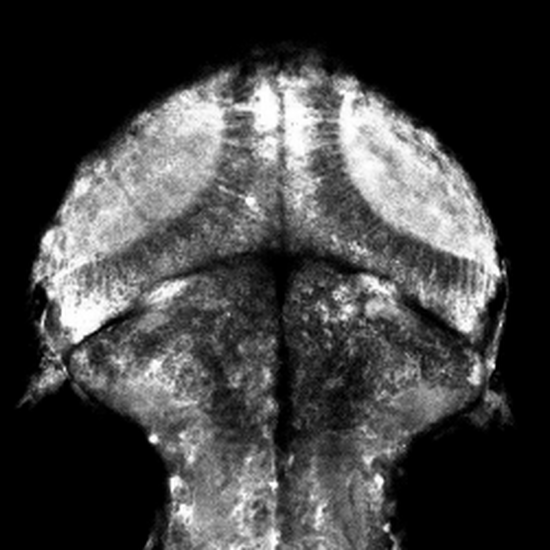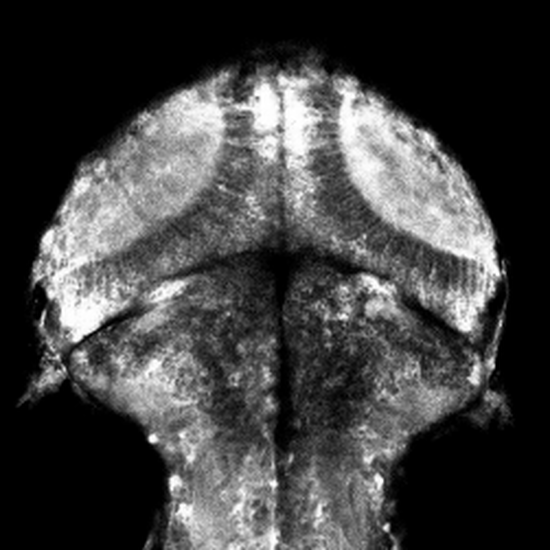
Non-Telecentric 2P microscopy for 3D random access mesoscale imaging (nTCscope)
Ultra-low-cost, easily implemented and flexible two-photon scanning microscopy modification offering a several-fold expanded three-dimensional field of view that also maintains single-cell resolution. Application of our system for imaging neuronal activity has been demonstrated on mice, zebrafish and fruit flies

Addgene’s AAV Data Hub
World Wide Series Seminar AAV are versatile tools used by neuroscientists for expression and manipulation of neurons. Many scientists have benefited from the high-quality, ready-to-use AAV prep service from Addgene, a nonprofit plasmid repository.
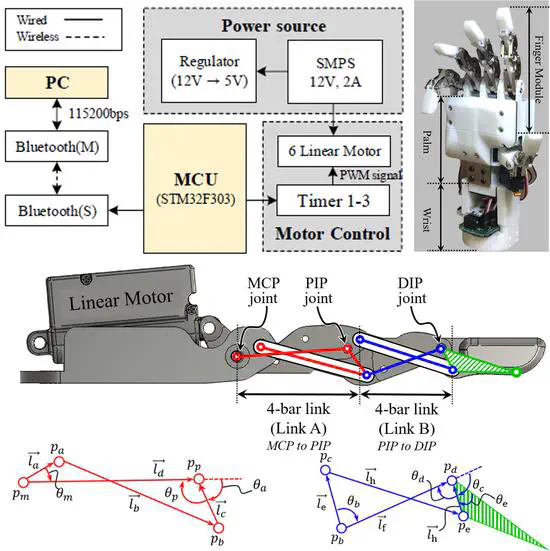
An Open-source Anthropomorphic Robot Hand System: HRI Hand
We present an open-source anthropomorphic robot hand system called HRI hand. Our robot hand system was developed with a focus on the end-effector role of the collaborative robot manipulator. HRI hand is a research platform that can be built at a lower price (approximately $500, using only 3D printing) than commercial end-effectors.
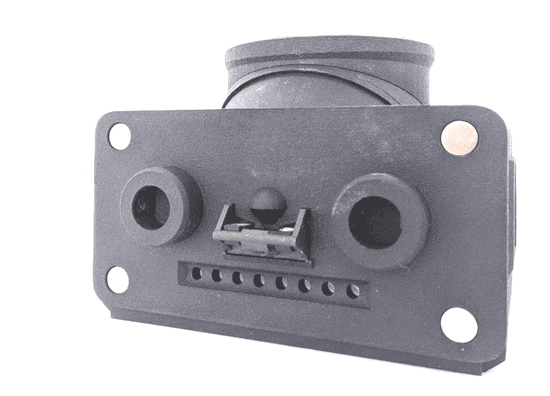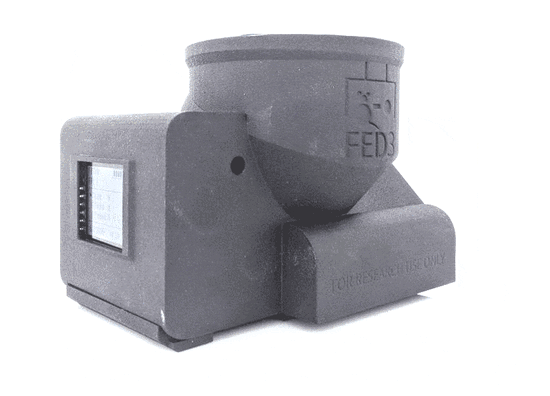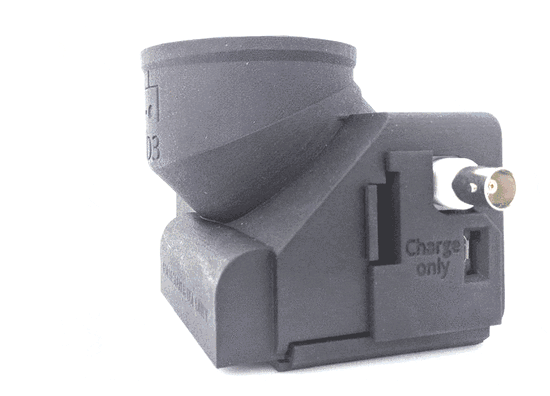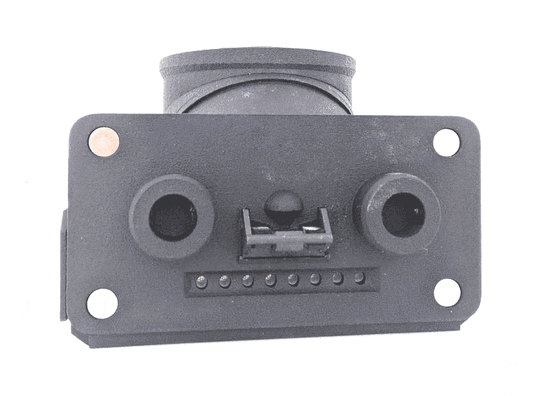
Feeding Experimentation Device ver3 (FED3)
World Wide Series Seminar FED3 is an open-source battery-powered device for home-cage training of mice in operant tasks. FED3 can be 3D printed and the control code is open-source and can be modified.
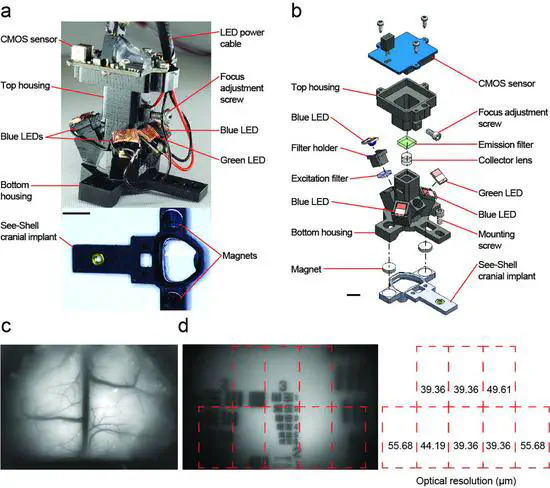
Head-Mounted Mesoscope
The advent of genetically encoded calcium indicators, along with surgical preparations such as thinned skulls or refractive index matched skulls, have enabled mesoscale cortical activity imaging in head-fixed mice. Such imaging studies have revealed complex patterns of coordinated activity across the cortex during spontaneous behaviors, goal-directed behavior, locomotion, motor learning,and perceptual decision making.
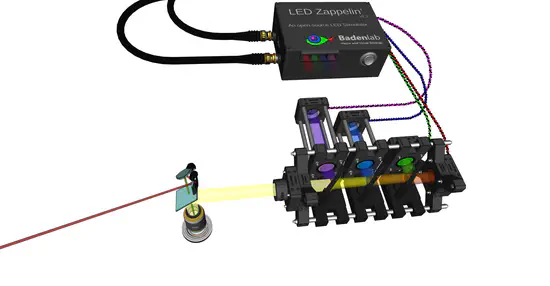
LED Zappelin'
Two-photon (2P) microscopy is a cornerstone technique in neuroscience research. However, combining 2P imaging with spectrally arbitrary light stimulation can be challenging due to crosstalk between stimulation light and fluorescence detection.

Suite2P
World Wide Series Seminar Suite2P is a very modular imaging processing pipeline written in Python which allows you to perform registration of raw data movies, automatic cell detection, extraction of calcium traces and infers spike times.
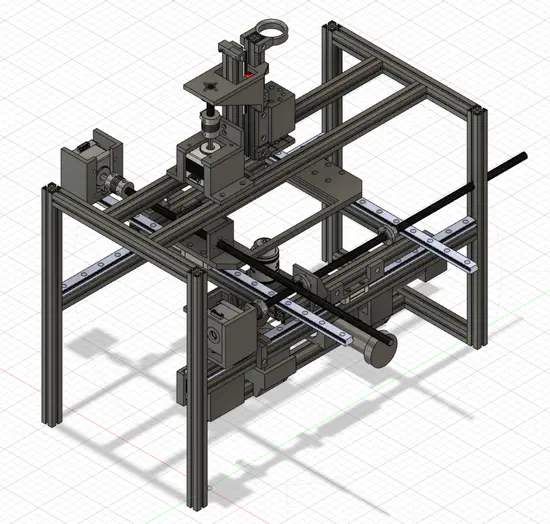
An open-source experimental framework for automation of high-throughput cell biology experiments
Modern Biology methods require a large number of high quality experiments to be conducted, which requires a high degree of automation. Our solution is an open-source hardware that allows for automatic high-throughput generation of large amounts of cell biology data.
BigPint Bioconductor package that makes BIG (RNA seq) data pint sized
The BigPint package can help examine any large multivariate dataset. However, we note that the example datasets and example code in this package consider RNA-sequencing datasets. If you are using this software for RNA-sequencing data, then it can help you confirm that the variability between your treatment groups is larger than that between your replicates and determine how various normalization techniques in popular RNA-sequencing analysis packages (such as edgeR, DESeq2, and limma) affect your dataset.

Collaborative Informatics and Neuroimaging Suite Toolkit for Anonymous Computation (COINSTAC)
COINSTAC provides a platform to analyze data stored locally across multiple organizations without the need for pooling the data at any point during the analysis. It is intended to be an ultimate one-stop shop by which researchers can build any statistical or machine learning model collaboratively in a decentralized fashion.
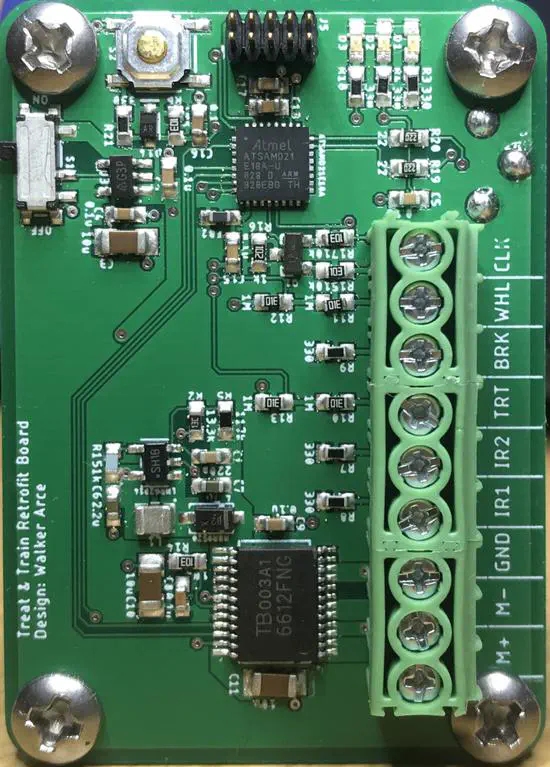
Computer-controlled dog treat dispenser
When performing canine operant conditioning studies, the delivery of the reward can be a limiting factor of the study. While there are a few commercially available options for automatically delivering rewards, they generally require manual input, such as using a remote control, in accordance with the experiment script.
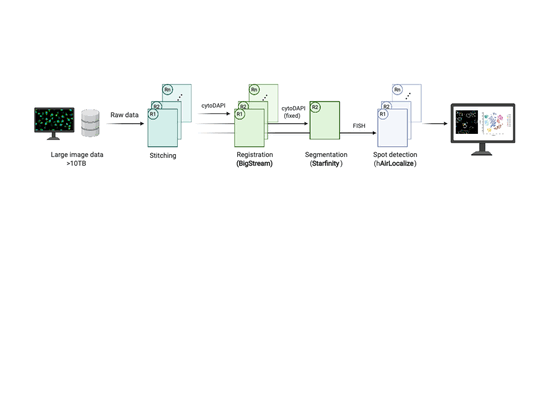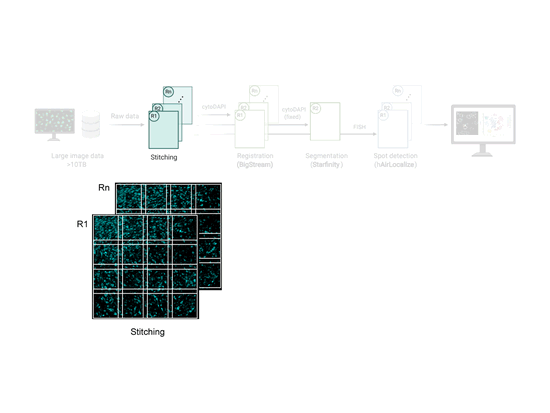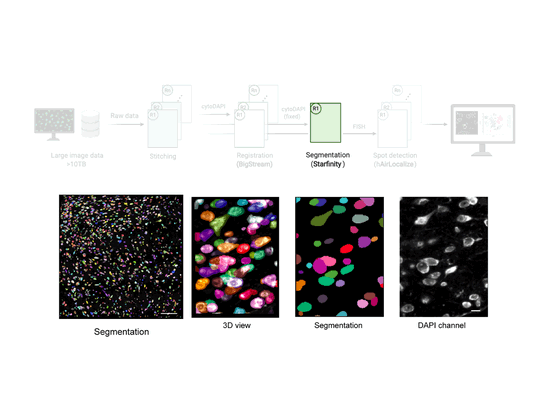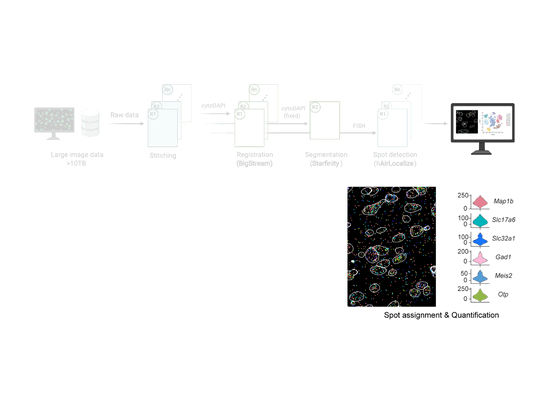
EASI-FISH
This workflow is used to analyze large-scale, multi-round, high-resolution image data acquired using EASI-FISH (Expansion-Assisted Iterative Fluorescence In Situ Hybridization). It takes advantage of the n5 filesystem to allow for rapid and parallel data reading and writing.
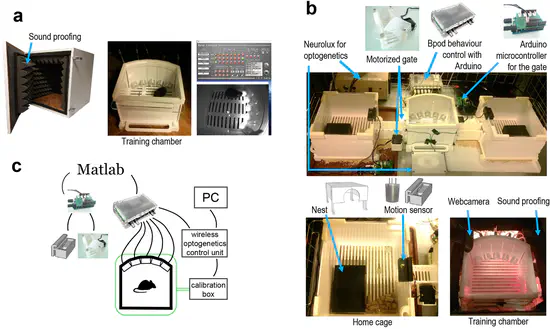
Efficient training of mice on the 5-choice serial reaction time task in an automated rodent training system
Experiments aiming to understand sensory-motor systems, cognition and behavior necessitate training animals to perform complex tasks. Traditional training protocols require lab personnel to move the animals between home cages and training chambers, to start and end training sessions, and in some cases, to hand-control each training trial.

Heuristic Spike Sorting Tuner (HSST)
Extracellular microelectrodes frequently record neural activity from more than one neuron in the vicinity of the electrode. The process of labeling each recorded spike waveform with the identity of its source neuron is called spike sorting and is often approached from an abstracted statistical perspective.

lab.js
lab.js makes online experimentation easy. It’s a free, open, online study builder and data collection framework for the behavioral, neuro- and cognitive sciences. Studies built with lab.js run in the laboratory as well as online, across a wide variety of data collection platforms, and provide excellent performance for all modes of stimulus presentation.

MNE-Python
MNE-Python is an open-source Python module for neuroscience data analysis. It implements many neuroscience-specific algorithms and statistical tools; has rich visualization capabilities that are both interactive and fully scriptable (for reproducibility); and integrates easily with general-purpose libraries like SciPy, Scikit-learn, and TensorFlow.
napari
Napari is a fast, interactive, multi-dimensional image viewer for Python. It’s designed for browsing, annotating, and analyzing large multi-dimensional images. It’s built on top of Qt (for the GUI), vispy (for performant GPU-based rendering), and the scientific Python stack (numpy, scipy).
openEyeTrack - An open source high-speed eyetracker
Vision is one of the primary senses, and tracking eye gaze can offer insight into the cues that affect decision-making behavior. Thus, to study decision-making and other cognitive processes, it is fundamentally necessary to track eye position accurately.
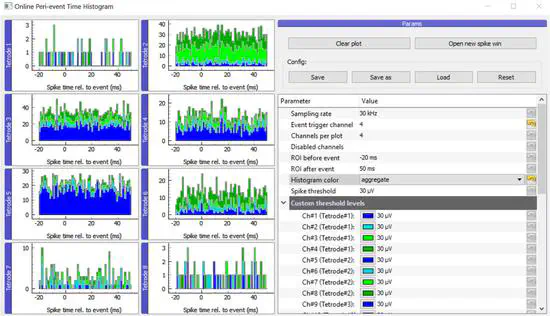
OPETH: Open Source Solution for Real-Time Peri-Event Time Histogram Based on Open Ephys
Single cell electrophysiology remains one of the most widely used approaches of systems neuroscience. Decisions made by the experimenter during electrophysiology recording largely determine recording quality, duration of the project and value of the collected data.
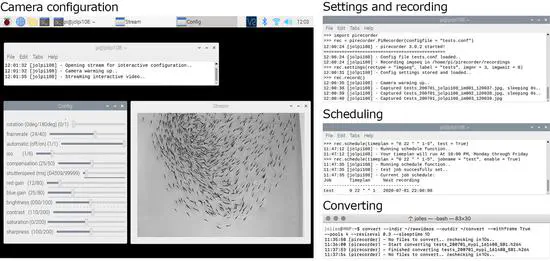
Pirecorder: Controlled and automated image and video recording with the raspberry pi
pirecorder is a Python package, built on the picamera and OpenCV libraries, that provides a flexible solution for the collection of consistent image and video data with the raspberry pi. It was developed to overcome the need for a complete solution to help researchers, especially those with limited coding skills, to easily set up and configure their raspberry pi to run large numbers of controlled and automated image and video recordings using optimal settings.

PocketPCR-Pocket size USB powered PCR Thermo Cycler
The PocketPCR is a so called thermocycler used to activate biological reactions. To do so the device raises and lowers the temperature of the liquid in the small tubes. The polymerase chain reaction (PCR) is a method widely used in molecular biology to make copies of a specific DNA segment.

CLIJ2
CLIJ2 is a GPU-accelerated image processing library for ImageJ/Fiji, Icy, Matlab and Java based on OpenCL. It comes with hundreds of operations for filtering, binarizing, labeling, measuring in images, projections, transformations and mathematical operations for images.
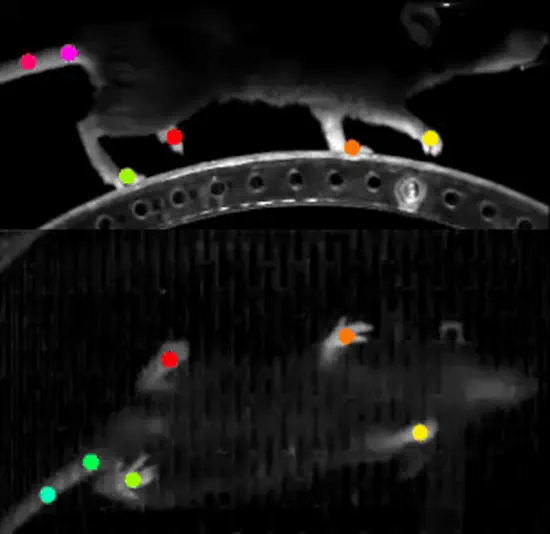
KineMouse Wheel
Who says you can’t reinvent the wheel?! This running wheel for head-fixed mice allows 3D reconstruction of body kinematics using a single camera and DeepLabCut (or similar) software. A lightweight, transparent polycarbonate floor and a mirror mounted on the inside allow two views to be captured simultaneously.
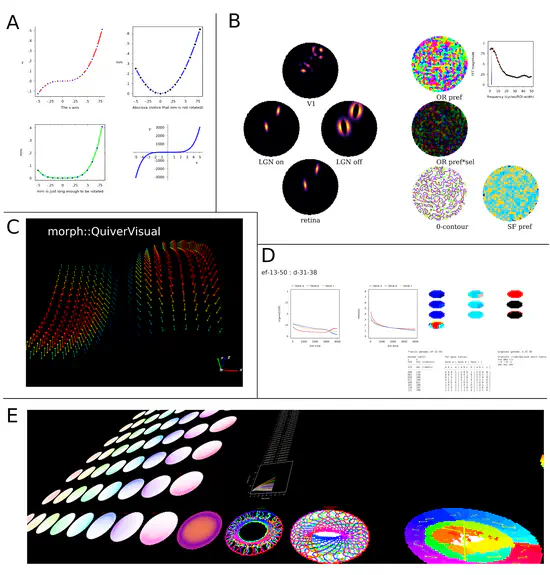
morphologica
morphologica is a header-only C++ library which provides simulation support facilities for simulations of dynamical systems. It helps with: Configuration: morphologica allows you to easily set up a simulation parameter configuration system, using the JSON reading and writing abilities of morph::Config.
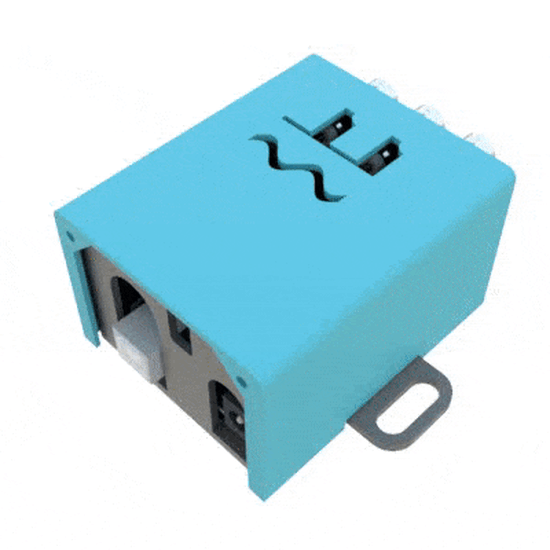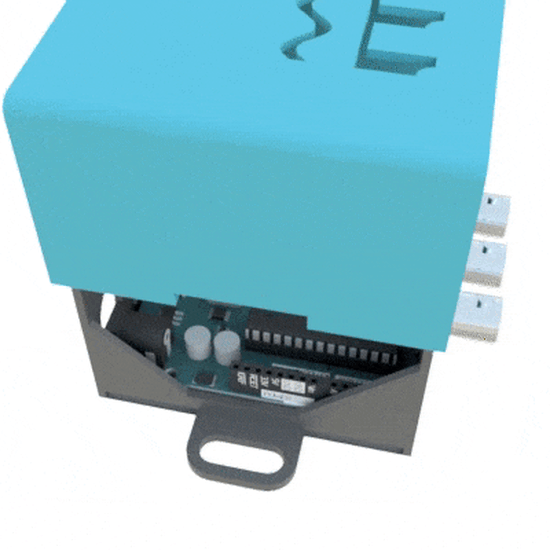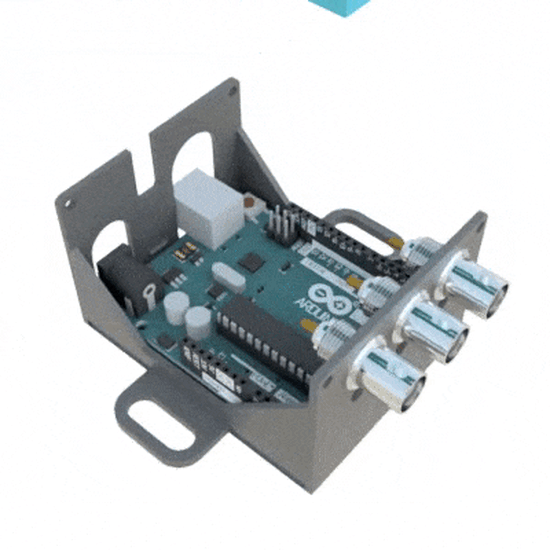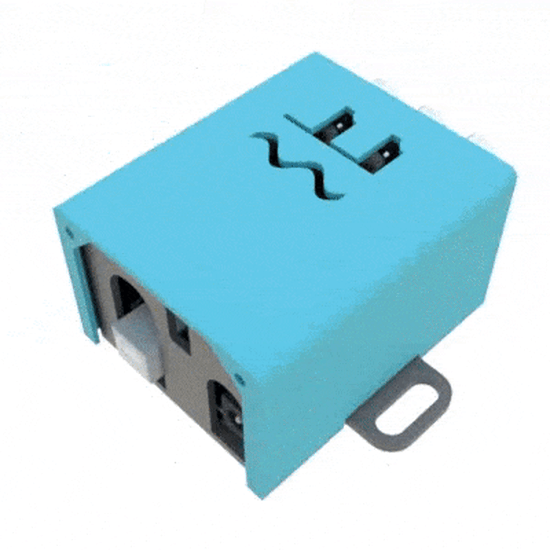
SignalBuddy
SignalBuddy is an easy-to-make, easy-to-use signal generator for scientific applications. Making friends is hard, but making SignalBuddy is easy. All you need is an Arduino Uno! SignalBuddy replaces more complicated and (much) more expensive signal generators in laboratory settings where one millisecond resolution is sufficient.

jsPsych
jsPsych is a JavaScript library for running behavioral experiments in a web browser. The library provides a flexible framework for building a wide range of laboratory-like experiments that can be run online.

NetPyNE
NetPyNE (Networks using Python and NEURON) is a Python package to facilitate the development, simulation, parallelization, analysis, and optimization of biophysical neuronal networks using the NEURON simulator. For more details, installation instructions, documentation, tutorials, forums, videos and more, please visit: www.
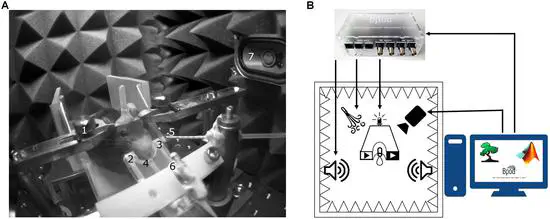
Open Source Tools for Temporally Controlled Rodent Behavior Suitable for Electrophysiology and Optogenetic Manipulations
Understanding how the brain controls behavior requires observing and manipulating neural activity in awake behaving animals. Neuronal firing is timed at millisecond precision. Therefore, to decipher temporal coding, it is necessary to monitor and control animal behavior at the same level of temporal accuracy.

Psygo
The easiest way to get started creating online behavioural experiments! psygo is a CLI tool that streamlines the development of custom jsPsych plugins, allowing you to create custom behavioural experiments that can be run online.

Brain Explained
Brain Explained explores the latest neuroscience that is fresh of the press. Tune in each month to listen to a deep dive interview into a research paper or thesis with the researcher.
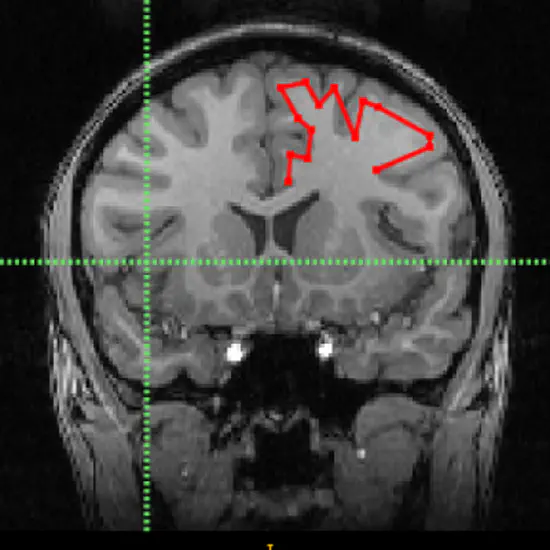
Greedy
Greedy is a tool for fast medical image registration. It is a fast CPU-based deformable image registration tool that can be used in applications where many images have to be registered in parallel.

ERPLAB
ERPLAB Toolbox is a free, open-source Matlab package for analyzing ERP data. It is tightly integrated with EEGLAB Toolbox, extending EEGLAB’s capabilities to provide robust, industrial-strength tools for ERP processing, visualization, and analysis.

TGAC Browser
The TGAC Browser is a Genomic Browser with novel rendering and annotation capabilities designed to overcome some shortcomings in available approaches. It was developed to visualize genome annotations from Ensembl Database Schema.
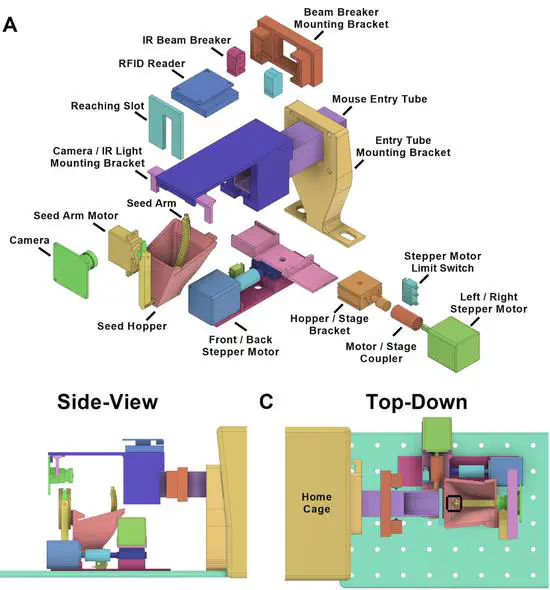
The home cage automated skilled reaching apparatus HASRA individualized training of group housed mice in a single pellet reaching task
We developed an automated apparatus and single pellet reach training paradigm for group housed mice within the home cage. This task allows individualized training progression and handedness of presentation for each mouse, minimizes stress induced by experimenter interaction, and enables experiments and data collection on a massive scale, previously impossible due to the demands of one-on-one behavioral testing.

neurolib
Easy whole-brain modeling for computational neuroscientists 👩🏿🔬💻🧠 In its essence, neurolib is a computational framework for simulating coupled neural mass models written in Python. It helps you to easily load structural brain scan data to construct brain networks where each node is a neural mass representing a single brain area.
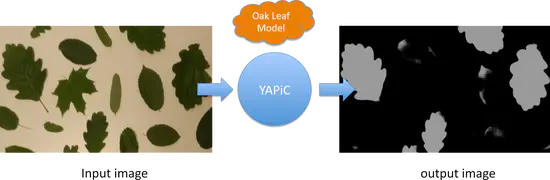
YAPiC
World Wide Series Seminar With YAPiC you can make your own customized filter (also called model or classifier) to enhance a certain structure of your choice with a simple Python based command line interface, installable with pip.

Computational Cognitive Neuroscience 4th Ed
This is the 4th edition of the online, freely available textbook, providing a complete, self-contained introduction to the field of Computational Cognitive Neuroscience, where computer models of the brain are used to understand a wide range of cognitive functions, including perception, attention, motor control, learning, memory, language, and executive function.

Emergent Neural Network Simulation Software
Neural network simulation software written in Go and Python, for developing biologically-based but also computationally functional neural models. Features an interactive 3D interface for visualizing networks and data, and has many implemented models of a wide range of cognitive phenomena.

Uncertainpy A python toolbox for uncertainty quantification and sensitivity analysis tailored towards computational neuroscience
Uncertainpy is a python toolbox for uncertainty quantification and sensitivity analysis tailored towards computational neuroscience. Uncertainpy is model independent and treats the model as a black box where the model can be left unchanged.
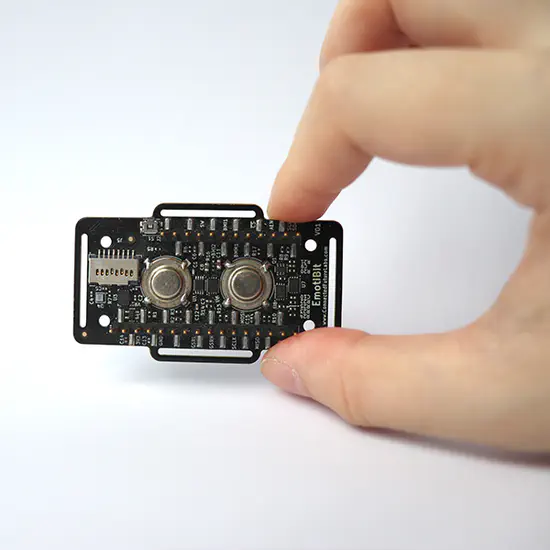
EmotiBit
EmotiBit is a wearable sensor to capture high-quality emotional, physiological, and movement data from just about anywhere on the body. Project Author(s) Sean Montgomery Project Links https://www.emotibit.com/ Project Video https://www.youtube.com/watch?v=jbcL2jzyWj4&ab_channel=EmotiBit

BrainSMASH
BrainSMASH is a Python-based framework for quantifying the significance of a brain map’s spatial topography in studies of large-scale brain organization. BrainSMASH was designed to generate synthetic brain maps with spatial autocorrelation (SA) matched to the SA of a target brain map.
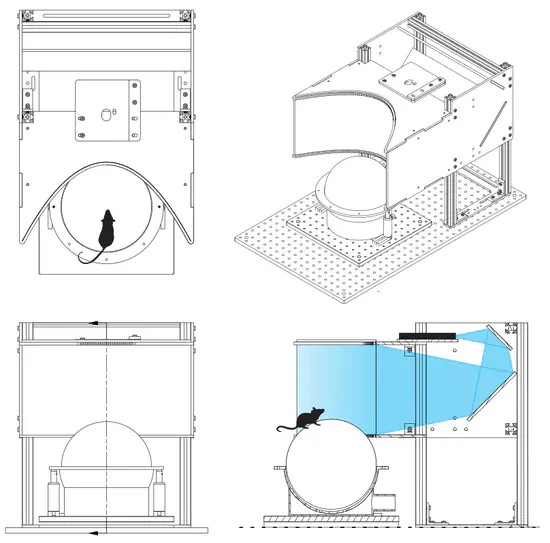
Mouse VR
Harvey Lab miniaturized mouse VR rig for head-fixed virtual navigation and decision-making tasks. The VR setup is comprised of several independent assemblies: The screen assembly: a laser projector projects onto a parabolic screen surrounding the mouse.

PsychRNN: An Accessible and Flexible Python Package for Training Recurrent Neural Network Models on Cognitive Tasks
PsychRNN is designed for neuroscientists and psychologists who are interested in RNNs as models of cognitive function in the brain. Despite growing interest in RNNs as models of brain function, this approach poses relatively high barriers to entry to researchers, due to the technical know-how required for specialized deep learning software (e.
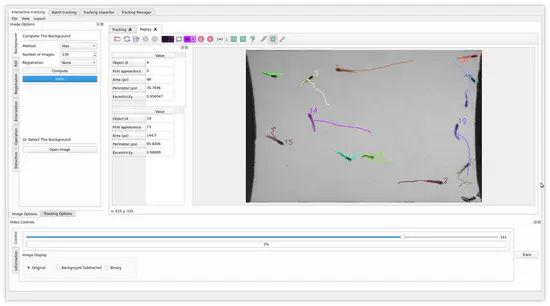
FastTrack
FastTrack is an open-source cross-platform tracking software. Easy to install and easy to use, it can track a large variety of systems from active particles to animals, with a known or unknown number of objects.
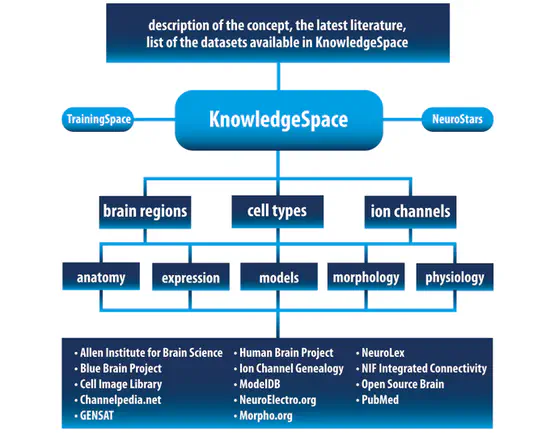
INCF KnowledgeSpace
KnowledgeSpace aims to be a globally-used, community-based, data-driven encyclopedia for neuroscience that links brain research concepts to data, models, and the literature that support them. Further it aims to serve as a framework where large-scale neuroscience projects can expose their data to the neuroscience community-at-large.

INCF NeuroStars
Neurostars is an open source question and answer site that serves the INCF network and the global neuroscience community as a forum for knowledge exchange between neuroscience researchers at all levels of expertise, software developers, and infrastructure providers.

INCF Portal
The INCF portal is the guide to the INCF activities and its community resources. INCF advances data reuse and reproducibility in brain research by coordinating the development of Open, FAIR, and Citable tools and resources for neuroscience
INCF TrainingSpace
TrainingSpace is an online hub that aims to make neuroscience educational materials more accessible to the global neuroscience community developed by the Training and Education Committee composed of members from the INCF network, HBP, SfN, FENS, IBRO, IEEE, BD2K, and iNeuro Initiative.

Open Source Brain
Open Source Brain, a platform for sharing, viewing, analyzing, and simulating standardized models from different brain regions and species. Model structure and parameters can be automatically visualized and their dynamical properties explored through browser-based simulations.

MorphoPy: A python package for feature extraction of neural morphologies
MorphoPy is an open software package written in Python3 that allows for visualization and processing of morphological reconstructions of neural data. It has been created to facilitate the translation from morphology graphs into descriptive features like density maps, morphometric statistics, and persistence diagrams for down-stream exploration and statistical analysis.

OpenCitations
OpenCitations is an independent infrastructure organization for open scholarship dedicated to the publication of open bibliographic and citation data by the use of Semantic Web (Linked Data) technologies. It is also engaged in advocacy for open citations, particularly by organizing the Workshops for Open Citations and Scholarly Metadata, and in its role as a key founding member of the Initiative for Open Citations (I4OC) and the Initiative for Open Abstracts (I4OA).

DIPY
DIPY is the paragon 3D/4D+ imaging library in Python. Contains generic methods for spatial normalization, signal processing, machine learning, statistical analysis and visualization of medical images. Additionally, it contains specialized methods for computational anatomy including diffusion, perfusion and structural imaging.

NeuroFedora
NeuroFedora is an initiative to provide a ready to use Fedora Linux based Free/Open source software platform for neuroscience. We believe that similar to Free software, science should be free for all to use, share, modify, and study.

BrainGlobe atlas API
Many excellent brain atlases exist for different species. Some of them have an API (application programming interface) to allow users to interact with the data programmatically (e.g. the excellent Allen Mouse Brain Atlas), but many do not, and there is no consistent way to process data from multiple sources.

culture_shock
Culture Shock is an open-source electroporator that was developed through internet based collaboration, starting on the DIYbio Google Group. It is an evolution on the traditional capacitive discharge circuit topology, instead using pulsed induction to enable a programmable waveform as well as reduce the size, weight, and cost of the equipment.
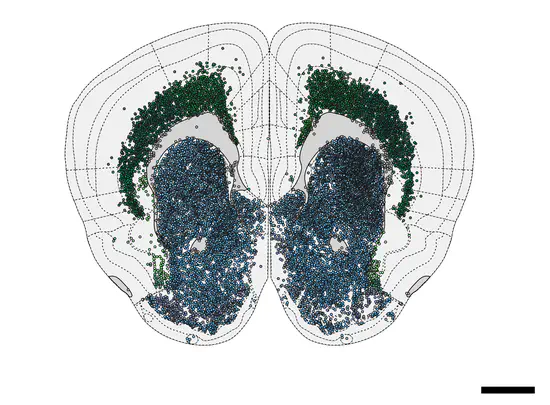
WholeBrain
WholeBrain is a software to create anatomical maps. With a code base in C/C++ with wrappers to R and JavaScript/WASM. The purpose of WholeBrain is to provide a user-friendly and efficient way for scientist with minimal knowledge of computers to create anatomical maps and integrate this information with behavioral and physiological data for sharing on the web.
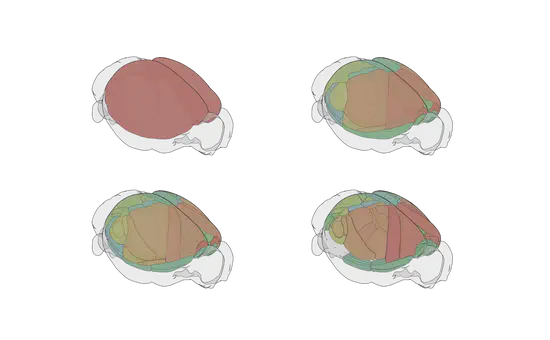
brainrender
brainrender is a python package for the visualization of three dimensional neuro-anatomical data. It can be used to render data from publicly available data set (e.g. Allen Brain atlas) as well as user generated experimental data.
JASP
JASP is a cross-platform statistical software program with a state-of-the-art graphical user interface. The JASP interface allows you to conduct statistical analyses in seconds, and without having to learn programming or risking a programming mistake.
PiDose
PiDose is an open-source tool for scientists performing drug administration experiments with mice. It allows for automated daily oral dosing of mice over long time periods (weeks to months) without the need for experimenter interaction and handling.

pyPhotometry
pyPhotometry is system of open source, Python based, hardware and software for neuroscience fiber photometry data acquisition, consisting of an acquisition board and graphical user interface. pyPhotometry supports data aquisition from two analog and two digital inputs, and control of two LEDs via built in LED drivers with an adjustable 0-100mA output.

SLEAP
SLEAP (Social LEAP Estimates Animal Poses) is a multi-animal pose tracker based on deep learning. It is the successor of LEAP (Pereira et al., Nature Methods, 2019) and was designed to deal with the problem of tracking body landmarks of multiple freely interacting animals.

Neuroimaging Informatics Tools and Resources Collaboratory (NITRC)
NeuroImaging Tools & Resources Collaboratory is an award-winning free web-based resource that offers comprehensive information on an ever expanding scope of neuroinformatics software and data. Since debuting in 2007, NITRC has helped the neuroscience community make further discoveries using software and data produced from research that used to end up lost or disregarded.
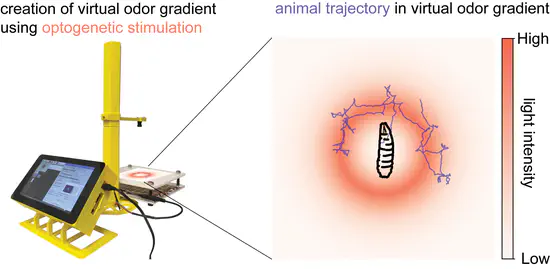
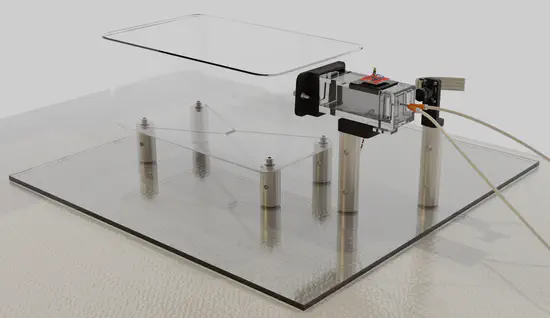
PiVR
World Wide Series Seminar PiVR is a system that allows experimenters to immerse small animals into virtual realities. The system tracks the position of the animal and presents light stimulation according to predefined rules, thus creating a virtual landscape in which the animal can behave.
ReproNim: A Center for Reproducible Neuroimaging Computation
ReproNim’s goal is to improve the reproducibility of neuroimaging science and extend the value of our national investment in neuroimaging research, while making the process easier and more efficient for investigators.

Simple Behavioral Analysis (SimBA)
Several excellent computational frameworks exist that enable high-throughput and consistent tracking of freely moving unmarked animals. SimBA introduce and distribute a plug-and play pipeline that enables users to use these pose-estimation approaches in combination with behavioral annotation for the generation of supervised machine-learning behavioral predictive classifiers.

Neurodata Without Borders
Neurodata Without Borders is a data standard for neurophysiology, providing neuroscientists with a common standard to share, archive, use, and build analysis tools for neurophysiology data. NWB is designed to store a variety of neurophysiology data, including data from intracellular and extracellular electrophysiology experiments, data from optical physiology experiments, and tracking and stimulus data.

OpenDrop Digital Microfluidics Platform
OpenDrop a modular, open source digital microfludics platform for research purposes. The device uses recent electro-wetting technology to control small droplets of liquids. Potential applications are lab on a chip devices for automating processes of digital biology.

Stytra
Stytra, a flexible, open-source software package, written in Python and designed to cover all the general requirements involved in larval zebrafish behavioral experiments. It provides timed stimulus presentation, interfacing with external devices and simultaneous real-time tracking of behavioral parameters such as position, orientation, tail and eye motion in both freely-swimming and head-restrained preparations.

Open Source Automated Western Blot Processor
Researchers in the biomedical area are always involved in methodologies comprising several processes that are repetitive and time-consuming; these researchers can take advantage of this time for other more important things.

OpenNeuro
A free and open platform for sharing MRI, MEG, EEG, iEEG, and ECoG data. With OpenNeuro, you can: Browse and explore public datasets and analyses from a wide range of global contributors.

Small cost efficient 3D printed peristaltic pumps
The project overall aim is to provide cost efficient solution to drive microfluidics systems for e.g. cell culture and organ on a chip applications. Pumps, valves and other accessories are ofter expensive to buy or very expensive to custom made.

Stackable titre plates for organ on a chip applications
Organ on a chip is typically difficult to achieve due to large technical challenges such as fabrication of chips and systems and biological challenges such as co-culture of cells. In this project we have developed a system to stack 12 well plates inserts on top of each other where each plate holds a tissue.
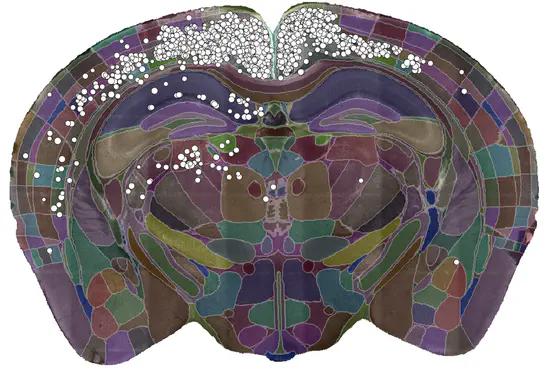
cellfinder
cellfinder is software from the Margrie Lab at the Sainsbury Wellcome Centre for automated 3D cell detection and registration of whole-brain images (e.g. serial two-photon or lightsheet imaging). It’s a work in progress, but cellfinder can:

DeepLabCut
DeepLabCut™ is an efficient method for 3D markerless pose estimation based on transfer learning with deep neural networks that achieves excellent results (i.e. you can match human labeling accuracy) with minimal training data (typically 50-200 frames).

Nilearn
Nilearn is a Python module for fast and easy statistical learning on NeuroImaging data. It leverages the scikit-learn Python toolbox for multivariate statistics with applications such as predictive modelling, classification, decoding, or connectivity analysis.

A Cartesian Coordinate Robot for Dispensing Fruit Fly Food
The fruit fly, Drosophila melanogaster, continues to be one of the most widely used model organisms in biomedical research. Though chosen for its ease of husbandry, maintaining large numbers of stocks of fruit flies, as done by many laboratories, is labour-intensive.
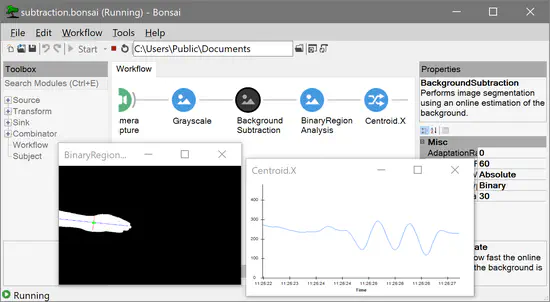
Bonsai
Bonsai is a high-performance, easy to use, and flexible visual programming language for designing closed-loop neuroscience experiments combining physiology and behaviour data. Bonsai has allowed scientists with no previous programming experience to quickly develop their own experimental rigs and is also being increasingly used as a platform to integrate new open-source hardware and software from the experimental neuroscience community.
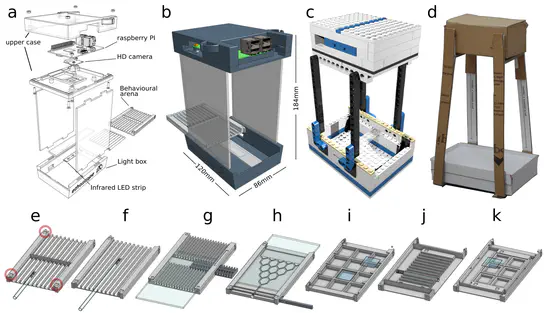
Ethoscopes
Ethoscopes are machines for high-throughput analysis of behavior in Drosophila and other animals. Ethoscopes provide a software and hardware solution that is reproducible and easily scalable. They perform, in real-time, tracking and profiling of behavior by using a supervised machine learning algorithm, are able to deliver behaviorally triggered stimuli to flies in a feedback-loop mode, and are highly customizable and open source.
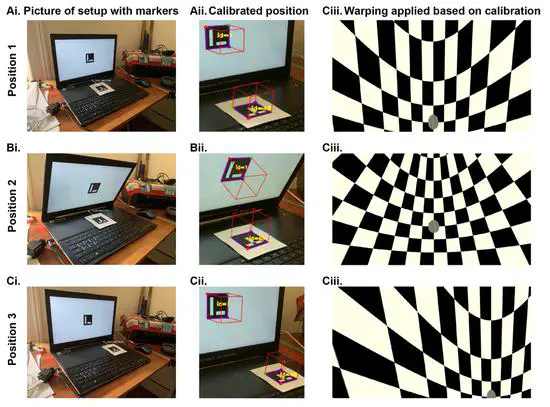
Bonvision
BonVision is an open-source closed-loop visual environment generator developed by the Saleem Lab and Solomon Lab at the UCL Institute of Behavioural Neuroscience in collaboration with NeuroGEARS. BonVision’s key features include:

Poseidon
The Poseidon is an open-source syringe pump and microscope system. It uses 3D printed parts and common components that can be easily purchased. It can be used in microfluidics experiments or other applications.
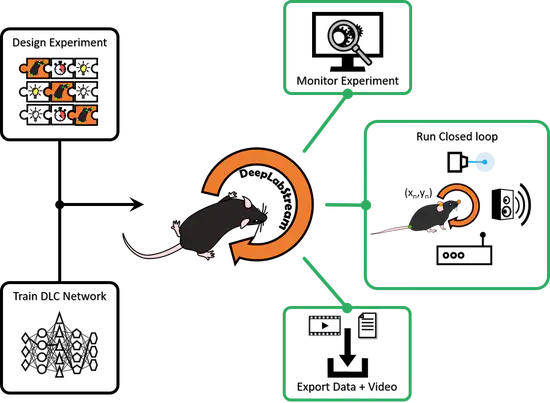
DeepLabStream
DeepLabStream is a python based multi-purpose tool that enables the realtime tracking of animals and manipulation of experiments. Our toolbox is adapted from the previously published DeepLabCut (Mathis et al., 2018) and expands on its core capabilities.

SpikeInterface
World Wide Series Seminar SpikeInterface is a unified Python framework for spike sorting. With its high-level API, it is designed to be accessible and easy to use, allowing users to build full analysis pipelines for spike sorting (reading-writing (IO) / preprocessing / spike sorting / postprocessing / validation / curation / comparison / visualization) with a few lines of code.

FishCam
We describe the “FishCam”, a low-cost (500 USD) autonomous camera package to record videos and images underwater. The system is composed of easily accessible components and can be programmed to turn ON and OFF on customizable schedules.

neuTube
neuTube is an open source software for reconstructing neurons from fluorescence microscope images. It is easy to use and improves the efficiency of reconstructing neuron structures accurately. The framework combines 2D/3D visualization, semi-automated tracing algorithms, and flexible editing options that simplify the task of neuron reconstruction.

OpenTrons
Today, biologists spend too much time pipetting by hand. We think biologists should have robots to do pipetting for them. People doing science should be free of tedious benchwork and repetitive stress injuries.
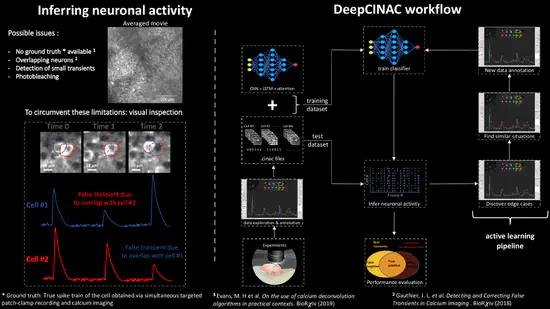
Deep Cinac
Two-photon calcium imaging is now widely used to infer neuronal dynamics from changes in fluorescence of an indicator. However, state of the art computational tools are not optimized for the reliable detection of fluorescence transients from highly synchronous neurons located in densely packed regions such as the CA1 pyramidal layer of the hippocampus during early postnatal stages of development.

Fingertip laser sensor
The fingertip laser project makes use of the sensor used in an Avago ADNS-9500 laser mouse, to improve the capabilities of robotic hands, giving them the capability to detect distance, surface type and slippage of grasped objects.

SciDraw
SciDraw is a free repository of high quality drawings of animals, scientific setups, and anything that might be useful for scientific presentations and posters. We want this repository to be as open as possible, so do not require signing up to post a drawing.
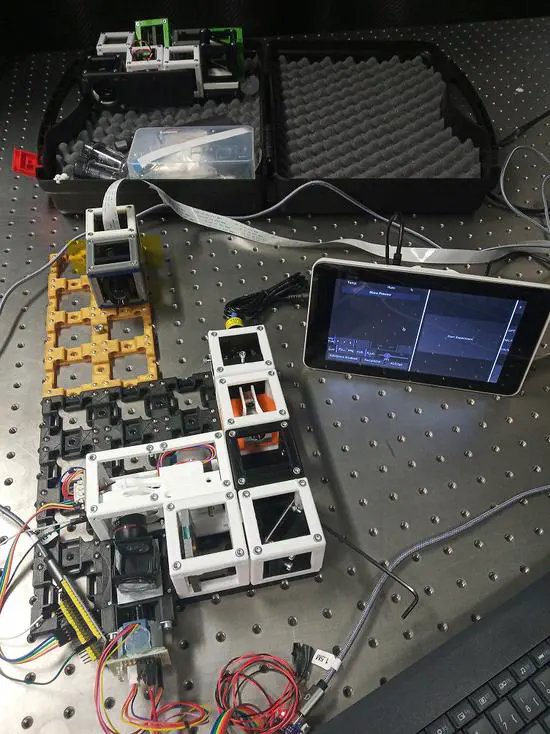
UC2
The open-source optical toolbox UC2 [YouSeeToo] simplifies the process of building optical setups, by combining 3D-printed cubes, each holding a specific component (e.g. lens, mirror) on a magnetic square-grid baseplate. The use of widely available consumables and 3D printing, together with documentation and software, offers an extremely low-cost and accessible alternative for both education and research areas.
Open Source Eye Tracking
The purpose of this project is to convey a location in 3 dimensional space to a machine, hands free and in real time. Currently it is very difficult to control machines without making the user provide input with their hands.
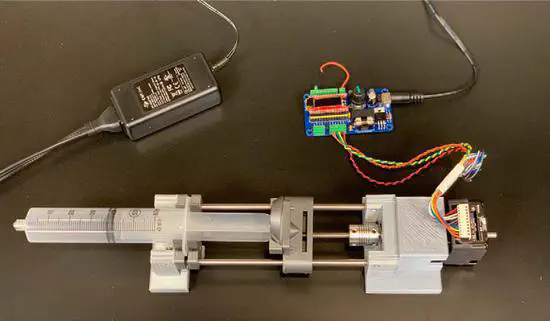
Open Source Syringe Pump Controller
Syringe pumps are a necessary piece of laboratory equipment that are used for fluid delivery in behavioral neuroscience laboratories. Many experiments provide rodents and primates with fluid rewards such as juice, water, or liquid sucrose.

Sample Rotator Mixer and Shaker
An open-source 3-D printable laboratory sample rotator mixer is developed here in two variants that allow users to opt for the level of functionality, cost saving and associated complexity needed in their laboratories.
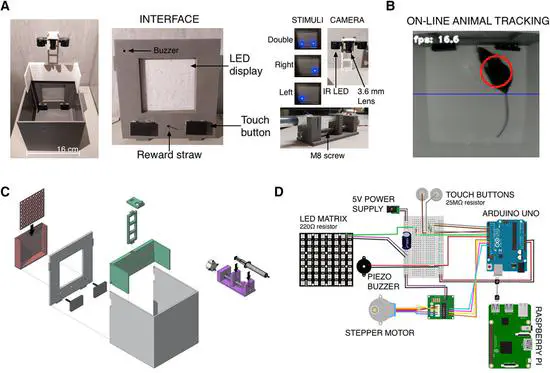
Automated Operant Conditioning
Operant conditioning (OC) is a classical paradigm and a standard technique used in experimental psychology in which animals learn to perform an action to achieve a reward. By using this paradigm, it is possible to extract learning curves and measure accurately reaction times (RTs).
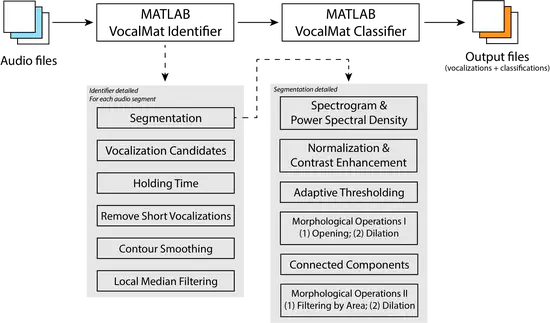
VocalMat
Mice emit ultrasonic vocalizations (USV) to transmit socially-relevant information. To detect and classify these USVs, here we describe the development of VocalMat. VocalMat is a software that uses image-processing and differential geometry approaches to detect USVs in audio files, eliminating the need for user-defined parameter tuning.

BossDB
BossDB is a volumetric database that lives in the AWS cloud. Hundreds of terabytes of electron microscopy, light microscopy, and x-ray tomography data are available for free download and study.

Neuroanatomy and Behaviour
Neuroanatomy and Behaviour (ISSN: 2652-1768) is a free open access journal for behavioural neuroscience and related fields. Powered by free open source software to eliminate costs and keep grant funds doing science.

3D Slicer
3D Slicer is a software for medical image informatics, image processing, and three-dimensional visualization. It’s extremely powerful and versatile with plenty of different options. It is a great tool for volume rendering, registration, interactive segmentation of images and even offers the possibility of running Python scripts thought an embedded Python interpreter.

Craniobot
The Craniobot is a cranial microsurgery platform that combines automated skull surface profiling with a computer numerical controlled (CNC) milling machine to perform a variety of cranial microsurgical procedures in mice.

Labmaker
LabMaker is a maker and assembly service for OPEN SCIENCE instruments. OPEN SCIENCE initiatives provide part lists or “Bill Of Materials” (BOM) for openly available scientific instruments. LabMaker bridges the gap between the BOM and the ready-to-use instrument for those not wanting to build by themselves.
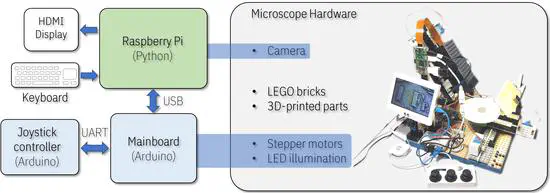
MicroscoPy
An open-source, motorized, and modular microscope built using LEGO bricks, Arduino, Raspberry Pi and 3D printing. The microscope uses a Raspberry Pi mini-computer with an 8MP camera to capture images and videos.

Colaboratory
Colaboratory is a free Jupyter notebook environment that runs in the cloud. Your notebooks get stored on Google Drive. The great advantage is that you don’t have to install anything (however, for some features you need a Google account) on your system to use it.
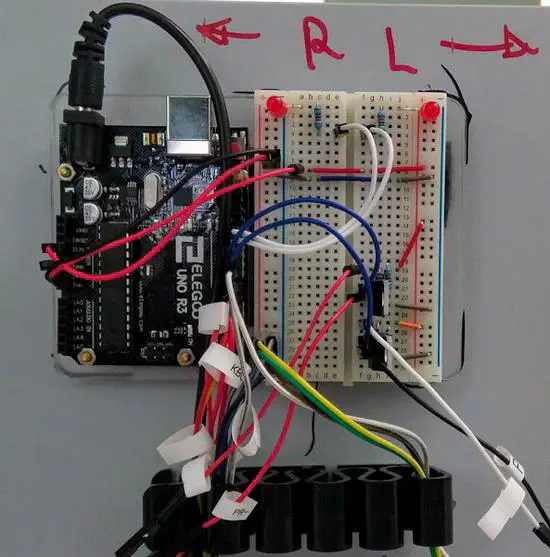
Autoreward 2
The motivation to start this project arises when we started to include a new behavioral paradigm in the lab, an alternation T-mace with return arms (like the one in Wood e_t al.

Backlog
Bellow is a list of interesting projects related to science and research, that we didn’t have time to curate yet. Feel free to browse through them and make comments and suggestions!

Computer clusters
Here are two projects that use card sized computers as the basic units for computing clusters: A 64 node cluster, build using pi’s and lego, built at the University of Southampton.

OpenFlexure
World Wide Series Seminar OpenFlexure is a 3D printed flexure translation stage, developed by a group at the Bath University. The stage is capable of sub-micron-scale motion, with very small drift over time.

OpenFuge
OpenFuge describes all the materials and gives step by step instructions to the assembly of a centrifuge that is able to deliver 6000 G’s of force and to rotate at 9000 RPM, while being able to hold 4 eppendorf tubes.
Psychophysics toolboxes
Roughly put, psychophysics studies the relationships of physical stimuli and their respective elicited sensations and perception. Psyhophysics also relates to the techniques used to probe these relationships and the toolboxes here presented are mainly dealing with these techniques.

Pulse Pal
Pulse Pal is an open and inexpensive (~$210) alternative to pulse generators used in neurophysiology research, and is most often used to create precisely timed light trains in optogenetics assays.

Python, NumPy, SciPy & Matplotlib
Python is a free programming language that is widely used, most of the software developed for Linux is written in Python. It contains several libraries that cover a lot of problem domains, from asynchronous processing to zip files.
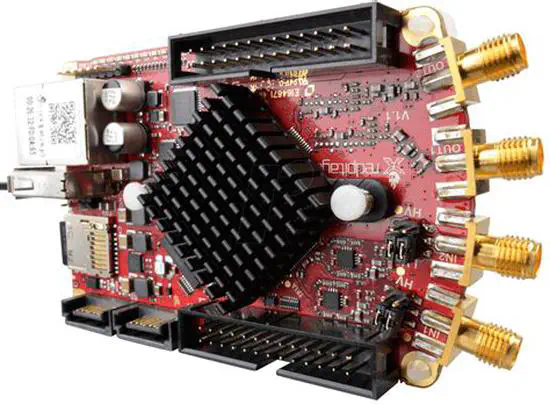
Red Pitaya
Red Pitaya is an computer+FPGA that has digital input and outputs and really fast analog inputs and outputs. It allows connection over ethernet and programming of custom routines. The system is powerful enough to have application in mostly all branches of neuroscience labs: oscilloscopes, signal generators and even a candidate for recording systems.

School of Data
School of Data is a global network that aims to train civil society in the practical use of the large amount of data available nowadays. The network is composed of individuals and organizations that carry out training programs, hands-on courses and other activities in different regions and countries of the world.

Spike Gadgets
A brief description of their current software (09.Sep.2016) is provided by one of their founders, Mattias Karlsson: State Script: Do you need to control lasers for optogenetics, stimulators, or other TTL-based devices with precise, temporally defined patterns?
Brainflow
BrainFlow BrainFlow is a library intended to obtain, parse and analyze EEG, EMG, ECG and other kinds of data from biosensors, it provides two APIs: Data Acquisition API to obtain data from BCI boards Signal Processing API which is completely independent and can be used without Data Acquisition API Both of these APIs are uniform for all supported boards, so it allows to write completely board agnostic code.

Genome RNAi
GenomeRNAi is a database containing phenotypes from RNA interference (RNAi) screens in Drosophila and Homo sapiens. In addition, the database provides an updated resource of RNAi reagents and their predicted quality.

GogoFuge
GogoFuge is a good example of the power of opensource designs. IT was based on the idea of the DremelFuge and altered to be a tabletop centrifuge with vortex capability. It was created by Keegan Cooke

Green Brain
The Green Brain project wants to create an artificial Apis mellifera brain and implement said brain into a robot, that will be able to fly and and behave just like a honey bee!
Image, Office suites, and other general purpose software
If you are using Linux, changes are that this page is not that useful for you, since most of these programs come installed by default. For you who are not yet into linux, most of these programs have Windows/Mac versions:
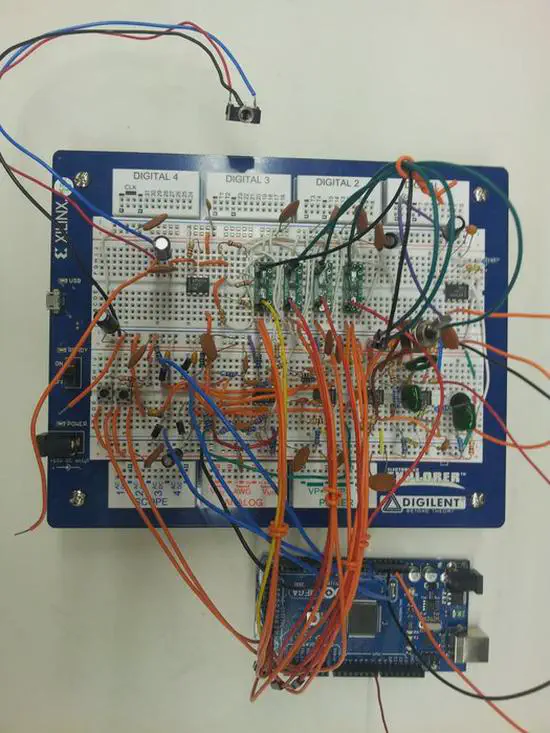
Intelligent hearing aid
Ojoshi at instructables.com has posted a manual on how to build this arduino based hearing aid system. From his instructables page: it has tuning functionality that allows the wearer to tune the amplification to his or her needs.
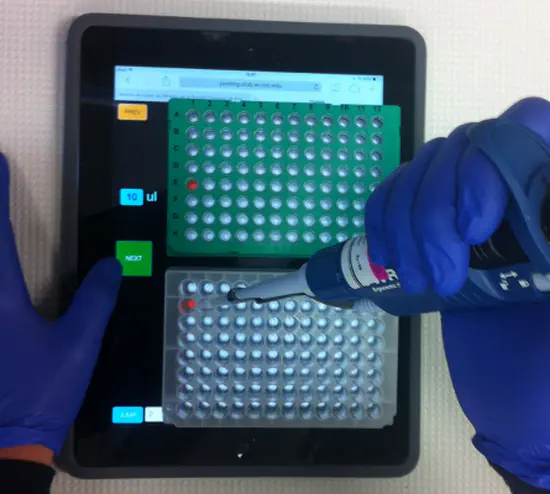
IPipet
IPipet is a neat system to help you not to lose track of which wells you have already pipetted in or from. The idea is simple, you place a tablet running a link with your specific pipetting protocol under your source and destination plates.
Lab management software
Since organisation of ideas, stocks, and projects is a major concern (or at least should be) of labs and researchers, here is a small compilation of cost free sofware to help out:

Micro-Manager
Micro-Manager is an ImageJ plugin dedicated to the control of microscopes. Their intent is to have a “one fits all” software for the control of microscopes, stages, filters and cameras. A comprehensive list of supported devives can be found on devices section of the project webpage.

Neuromorpho
NeuroMorpho.Org is a centrally curated inventory of digitally reconstructed neurons associated with peer-reviewed publications. It contains contributions from over 100 laboratories worldwide and is continuously updated as new morphological reconstructions are collected, published, and shared.
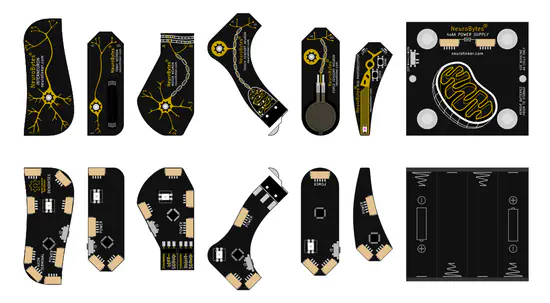
NeuroTinker
NeuroTinker project is all about hardware emulated neurons. The creators made them in a way that each hardware neuron has excitatory and inhibitory inputs and one output that can be split up to affect dowsntream neurons.
NiBabel
NiBabel is a python package, under the NiPy project, that aims at unifying the process of opening different medical and neuroimaging file formats, including: ANALYZE,GIFTI, NIfTI1, MINC, MGH and ECAT as well as PAR/REC.
Nipy
NiPy is an effort to make brain imaging research easier and more clear. This is implemented by providing a series of software that deal with file IO, analysis, and interfaces & pipelines.
Nose poke device for rats using arduino and 3d printed parts
This is a small set of instructions on how to build a nose poke device for rats, using an arduino, some 3D printed parts and some off-the-shelf electronic components. All the files necessary to reproduce this can be found here

Open Microscopy Environment
The Open Microscopy Environment is a collaborative project between several labs. They are developing file formats and software standards for light microscopy. Within the project they have BIO-formats, a Java library for reading and writing data.
Operating systems
Linux is an open source operating system and it is the major OS used in servers and supercomputers. Ubuntu, one of the best known distributions has been gaining space in the personal computing scene, now days already being factory shipped by major manufacturers.

Super-Releaser
Ever thought about making soft robots? The folks at Super-Releaser have, and they are doing very cool projects! Some for medical applications and some for research purposes. Check one of their cool robots below:

5 Dollar PCR machine
The 5 dollar PCR machine is a project from David Ng. he created a very interesting design for the PCR machine. Instead of using eppendorfs, he is using teflon tubes and three different heating elements, which allows for cheaper (he has a working PCR machine for 5 dollars!

Open bionics
The Open bionics project was inspired by the Yale open hand project, aiming to develop light, affordable, and modular robot hands and myoelectric prosthesis. Also they want to make them easy to replicate using off the shelf materials.
Open prosthetics and robotics
With the rise of low cost 3D printers, and other cheap manufacturing tools, the field of robotics and prosthetics has been gaining quite a few open source projects. Two very nice compilations can be found at openrobot hardware and at Soft robotics toolkit.

Backyard Brains
Backyard brains started out producing low cost, portable, electrophysiology systems to bring neuroscience to classrooms and help promote it. “Backyard brains wants to be for neuroscience, what the telescope is for astronomers” – meaning that the idea is that with a couple of hundred dollars anyone can get one of these recording systems and start doing experiments, like amateur astronomers can buy telescopes and start observing the cosmos.

10$ smartphone microscope
This neat little project uses some plexi-glass, lens extracted from a laser pointer to harvest the power of smartphone cameras for some very big amplifications! Yoshinok manged to see cell plasmolysis and some other cool features with it.

Addgene
Addgene is a non-profit company that makes the share of plasmids easier by making a plasmid database and linking them to the papers where they were described. In this way they take on the job of maintaining plasmids and shipping them to requesting scientists.
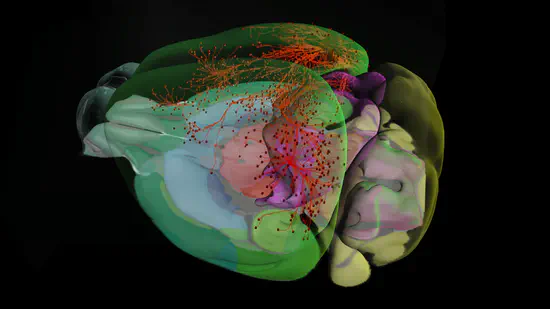
Allen Brain Map
The Brain Map is one of the initiatives of the Allen Institute. It is a data portal that encompasses different projects: the Allen Institute has created a set of large-scale programs to understand the fundamentals of the cortex.
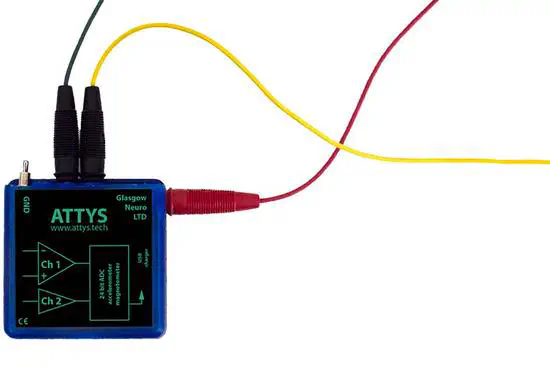
Attys
Attys is an wearable data acquisition device with a special focus on biomedical signals such as heart activity (ECG), muscle activity (EMG) and brain activity (EEG). It’s open firmware, open API and has open source applications on github in C++ and JAVA to encourage people to create their own custom versions for mobile devices, tablets and PC.
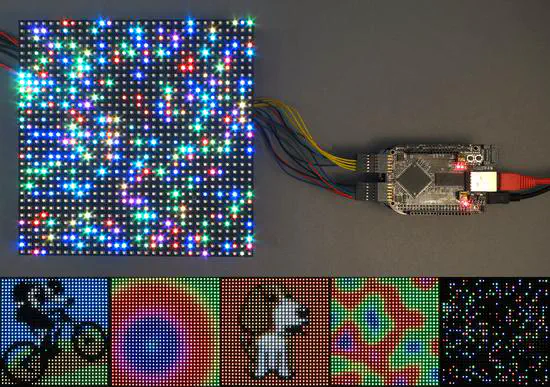
BB LED Matrix
This project uses a 32X32 LED array (1024 LEDs in total) and a beagle bone black board. The page describing the project has very nice explanations on how the whole system works (and LED displays in general).
Big Neuron
Big Neuron wants to create a standard for the field of single neuron reconstruction. Because the data available comes from different structures, different organisms, using different collection and analyses algorithms and is in the range of petabytes (according to the project site), there is a strong need for standards that will allow this huge amount of data to be compared.
BioAmp
BioAmp is a biopotential acquisition device (EEG, ECG, EMG, EOG, etc.) developed in the Prototyping Laboratory at the School of Engineering of the National University of Entre Rios (Argentina). Main features:

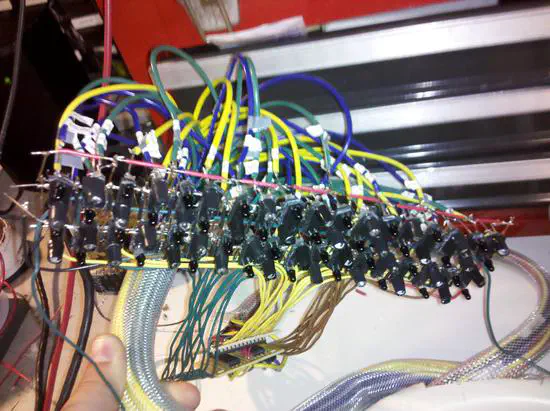
Blinkenschild
Blinkenschild is a portable sign consisting of 960 RGB LEDs. The images/movies to be displayed are stored in a SD card in a Teensy3 board and controlled via bluetooth. Resolution is not as high as LCD monitors but the refresh rate is much higher:

Boinc
Boinc is a platform for volunteer computing. Briefly, volunteer computer is a system where computer processor’s idle time (those periods where your computer is on, but not being used for anything) is turned into calculation time via a custom written software.
BPM Biosignal
BPM Biosignal is a two stage amplifier created mainly for educational purposes. Check their YouTube Channel.
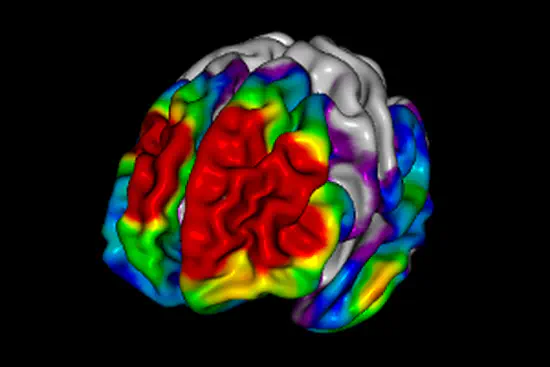
Brain Map
BrainMap expands the accessible DIY projects for brain activity measurements. This is the conclusion project of Patrick Dear and Mark Bunney Jr. at Cornell university where they used infrared leds to measure differences in blood flow at the scalp and map the motor cortex.
BrainBrowser
BrainBrowser is a collection of open source, web-based 3D data visualization tools, mainly for neuroimaging studies. It is built using open technologies such as WebGL and HTML5. It allows exploration of cortical surface models (MNI and Wavefront OBJ, as well as FreeSurfer ASCII surface format) and volumetric MINC data.
Computer Vision and motion tracking software
Motion tracking can be really useful in neurosciences, for automatic measurements of behaviour, among other things. Here you’ll find a small list of tracking softwares or libraries used to build such softwares:
Crowd funding
As many other things that are being decentralized with the advent of the internet, so is research. One of the very things being decentralized is the funding source for research projects.
Data repositories
In here are some examples of tools that can be used to share/store data collected. Published a paper and think that people would benefit from looking at the raw data? Want to make that data that has been stored for years useful?
DIY PCR
Katharina and Alex are developing a classic PCR machine: 16 samples and a heated lid. You can find more details of their project here Here is a demo video:
DremelFuge
DremelFuge is a very simple and clever centrifuge, buit perhaps not the safest one (be careful if you end up using it!). It takes advantage of 3d printing technology to print an adaptor that goes on to a Dremel (a precision tool that has really high rotation rates).
Fiji
Fiji is a distribution of ImageJ. The idea of the developers is to make the life of scientists easier by bundling ImageJ with nicely organised plugins and auto update function.
Interesting projects
It is great that there are other interesting projects out there that are also concerned with making science available to more people! Here is a short list of projects I came across.
NeuroElectro
NeuroElectro wants to extract information about neuron types, morphology, electrophysiology properties from papers, using text mining algorithms and gathers them in a database. Our goal is to facilitate the discovery of neuron-to-neuron relationships and better understand the role of functional diversity across neuron types.

Open BCI
OpenBCI is a complete open source EEG system that can be built either on top of an Arduino (8-bit system), or on top of chipKIT (32-bit system), which gives the system more local memory and allows for faster speeds.

Open EEG
The openEEG project aims at describing and putting manuals for building a two channel EEG system for about U$200. More on instructions on how to build one, can be found here.
Open Ephys
World Wide Series Seminar Open Ephys is a great initiative to create a suite that encompasses hardware for LFP and spiking recording, optogenetics combined with custom written software for microstimulation, environmental stimuli, extracellular recording and optogen.

Open ExG
OpenHardwareExG: is a project that provides both open source hardware and software for the measurement and analysis of different types of biosignals From the project page: About the OpenHardwareExG project Project goals The main goal of the project is to build a device that allows the creation of electrophysiologic signal processing applications.
Open lab notebooks
Open notebooks are opening up science in the very first steps, making records of ideas, plans that didn’t work and protocols that failed available publicly. This allows others to avoid trailing the same dead end roads, saving time, money and human power.

Open PCR
Open PCR is an open source PCR machine with heated lid and space for 12 samples
Open science framework
From the Open science framework webpage: The Open Science Framework (OSF) is part network of research materials, part version control system, and part collaboration software. The purpose of the software is to support the scientist’s workflow and help increase the alignment between scientific values and scientific practices.
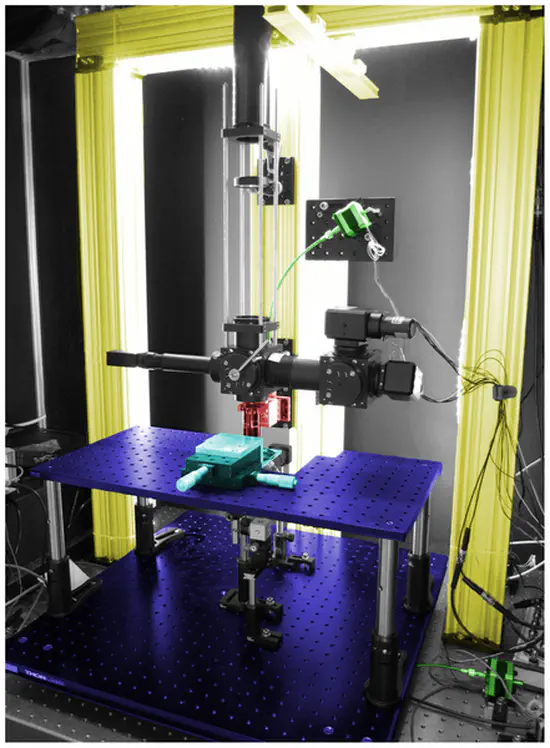
OpenStage
Open stage is a low-cost motorised microscope stage capable of movement in the micrometer range. It features manual control via a control-pad, different movement velocities and pc communication through the serial port.
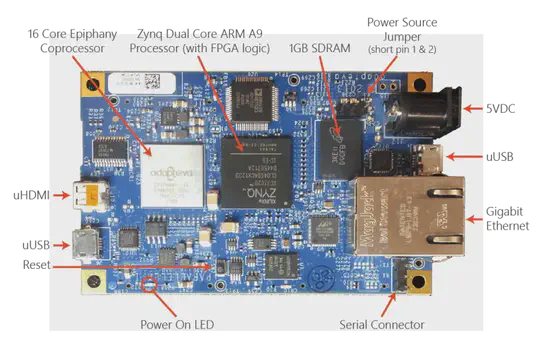
Parallela
Parallela, an open source, open access card sized supercomputer, has the mission of bringing parallel computing to the masses by combining multiple RISC processors and very low power consumption. Produced by the Adapteva company.
PySpace
PySpace is a signal processing and classificiation environment for Python. Modular software for processing of large data streams that has been specifically designed to enable distributed execution and empirical evaluation of signal processing chains.
Python for Neurosciences (Frontiers collection)
Frontiers has created not one but two nice collections about open source software for neurosciences written in Python. Here is collection 1 Here is collection 2 In these collections the readers will find a lot of nice resources, ranging from stimulus generation, to data formatting and analysis.
Signal Generators
Every lab needs a signal generator once in a while. They are useful to see if your acquisition program is working properly, to test why a certain piece of equipment is not working properly or to generate cues and targets at behavioural paradigms.
Simulations
Ever thought about playing with a virtual worm? or interacting with a simulated bee brain? Sounds interesting no? These are just two projects that offer anyone the opportunity to play around with brain/neuronal simulations and models.

Skinner Box with RPi+Python
This project was developed by Katherine Scott to be presented at the PyCon 2014. She developed a skinner box for her pet rats using a raspberry pi and some 3D printed parts.

Stereo microscope
Although stereo microscopes are an essential piece of hardware in biology labs, sometimes we wish they had more features, like the possibility to record the magnified images with a camera, or have a better lighting system to enhance contrast on those small samples.
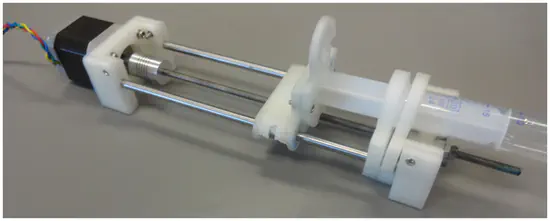
Syringe Pump
From the Pearce lab, this syringe pump was published in Plos One and is built using 3d printed parts, stepper motors and a raspberry pi, costing 5% or less than commercial available systems.
Takktile
Takktile, is a tactile sensor to be used on robotic applications. The developers want to make it move away from the closed walls of research institutions by making it open source and cheap.

tDCS
Although of simple complexity and using low currents, this tDCS machine is still to be considered a piece of equipment that could be dangerous both in the assembly and in the operation phases, so please inform yourself as best as you can before either of these steps!
Tensor Flow
Google has packaged their deeplearning machine learning tools and made it open source. The project is called tensorflow, and is available here. Some nice tutorials on the website, so that with a bit of patience, people can start to deep their toes into machine learning!
The Visible Human project
The Visible Human Project is a database of anatomical images (MR, CT and radiography) from male and female bodies. Information about the database is translated into a couple of different languages.

The Yale open hand project
The Yale open hand project, has a similar purpose of the open hand project, that is, to make prosthetic hands more widely available through the lowering of costs. They have a different design from the open hand project.
Vision Egg
Vision Egg is a Python library for generating visual stimuli. In more detail, it is a high level interface in between Python and OpenGL, and can use inexpensive consumer grade graphics cards to generate precise visual stimuli.
Web portals
Here are some open learning sources, they go from sites that interactively teach one how to code, to efforts in publishing free college textbooks. Khan Academy: Is composed of a series of lectures and exercises on a wide range of topics from basic multiplication to linear algebra and information theory.
YouTube as a resource for Open Science Hardware
When working with Open Hardware, Google search will become your friend, whether you want it or not. Other search engines, such as DuckDuckGo won’t cut it (it is much harder to find what you are looking for, especially in cases where you don’t know all specific terms).
Seminars
We run a seminar series where tool and methods can be showcased by their developers!

Trackoscope
World Wide Series Seminar Cells and microorganisms are motile, yet the stationary nature of conventional microscopes impedes comprehensive, long-term behavioral and biomechanical analysis. The limitations are twofold: a narrow focus permits high-resolution imaging but sacrifices the broader context of organism behavior, while a wider focus compromises microscopic detail.

Mobilefuge: Low-cost, Open-source 3D-printed centrifuge
We made a low-cost centrifuge that can be useful for carrying out low-cost LAMP based detection of SARS-Cov2 virus in saliva. The 3D printed centrifuge (Mobilefuge) is portable, robust, stable, safe, easy to build and operate.

The Virtual Macaque Brain
A whole-cortex macaque structural connectome constructed from a combination of axonal tract-tracing and diffusion-weighted imaging data. Created for modeling brain dynamics using TheVirtualBrain (thevirtualbrain.org) platform. A detailed description and example usage can be found in the paper here: https://www.

BrainGlobe
World Wide Series Seminar BrainGlobe is a suite of Python-based computational neuroanatomy software tools. We provide software packages for the analysis and visualisation of neuroanatomical data, particularly from whole-brain microscopy. In addition, we provide tools for working with brain atlases, to simplify development of new tools and aid collaboration and cooperation by adopting common standards.

Kilosort
World Wide Series Seminar Kilosort is a software package for identifying neurons and their spikes in extracellular electrophysiology, a process known as “spike sorting”. Kilosort has been primarily developed and tested on the Neuropixels 1.

Non-Telecentric 2P microscopy for 3D random access mesoscale imaging (nTCscope)
Ultra-low-cost, easily implemented and flexible two-photon scanning microscopy modification offering a several-fold expanded three-dimensional field of view that also maintains single-cell resolution. Application of our system for imaging neuronal activity has been demonstrated on mice, zebrafish and fruit flies

Addgene’s AAV Data Hub
World Wide Series Seminar AAV are versatile tools used by neuroscientists for expression and manipulation of neurons. Many scientists have benefited from the high-quality, ready-to-use AAV prep service from Addgene, a nonprofit plasmid repository.

Feeding Experimentation Device ver3 (FED3)
World Wide Series Seminar FED3 is an open-source battery-powered device for home-cage training of mice in operant tasks. FED3 can be 3D printed and the control code is open-source and can be modified.

Suite2P
World Wide Series Seminar Suite2P is a very modular imaging processing pipeline written in Python which allows you to perform registration of raw data movies, automatic cell detection, extraction of calcium traces and infers spike times.

An open-source experimental framework for automation of high-throughput cell biology experiments
Modern Biology methods require a large number of high quality experiments to be conducted, which requires a high degree of automation. Our solution is an open-source hardware that allows for automatic high-throughput generation of large amounts of cell biology data.

YAPiC
World Wide Series Seminar With YAPiC you can make your own customized filter (also called model or classifier) to enhance a certain structure of your choice with a simple Python based command line interface, installable with pip.

NeuroFedora
NeuroFedora is an initiative to provide a ready to use Fedora Linux based Free/Open source software platform for neuroscience. We believe that similar to Free software, science should be free for all to use, share, modify, and study.

PiVR
World Wide Series Seminar PiVR is a system that allows experimenters to immerse small animals into virtual realities. The system tracks the position of the animal and presents light stimulation according to predefined rules, thus creating a virtual landscape in which the animal can behave.

DeepLabStream
DeepLabStream is a python based multi-purpose tool that enables the realtime tracking of animals and manipulation of experiments. Our toolbox is adapted from the previously published DeepLabCut (Mathis et al., 2018) and expands on its core capabilities.

SpikeInterface
World Wide Series Seminar SpikeInterface is a unified Python framework for spike sorting. With its high-level API, it is designed to be accessible and easy to use, allowing users to build full analysis pipelines for spike sorting (reading-writing (IO) / preprocessing / spike sorting / postprocessing / validation / curation / comparison / visualization) with a few lines of code.

OpenFlexure
World Wide Series Seminar OpenFlexure is a 3D printed flexure translation stage, developed by a group at the Bath University. The stage is capable of sub-micron-scale motion, with very small drift over time.
Open Ephys
World Wide Series Seminar Open Ephys is a great initiative to create a suite that encompasses hardware for LFP and spiking recording, optogenetics combined with custom written software for microstimulation, environmental stimuli, extracellular recording and optogen.