Posts

The Poseidon is an open-source syringe pump and microscope system. It uses 3D printed parts and common components that can be easily purchased. It can be used in microfluidics experiments or other applications.
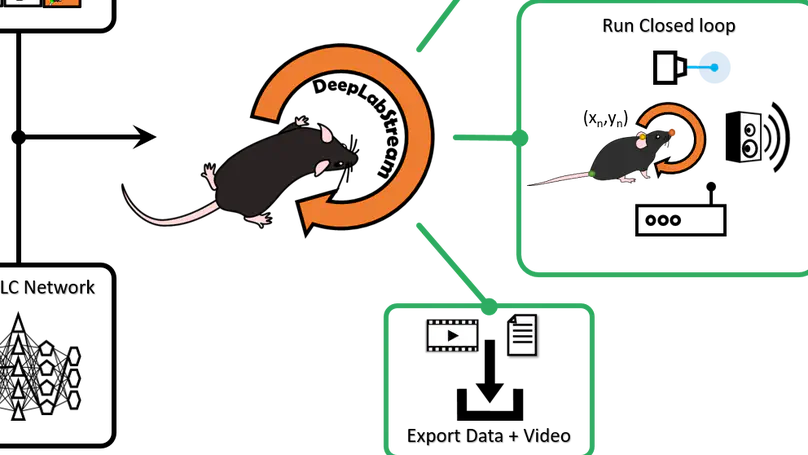
DeepLabStream is a python based multi-purpose tool that enables the realtime tracking of animals and manipulation of experiments. Our toolbox is adapted from the previously published DeepLabCut (Mathis et al., 2018) and expands on its core capabilities.
World Wide Series Seminar SpikeInterface is a unified Python framework for spike sorting. With its high-level API, it is designed to be accessible and easy to use, allowing users to build full analysis pipelines for spike sorting (reading-writing (IO) / preprocessing / spike sorting / postprocessing / validation / curation / comparison / visualization) with a few lines of code.
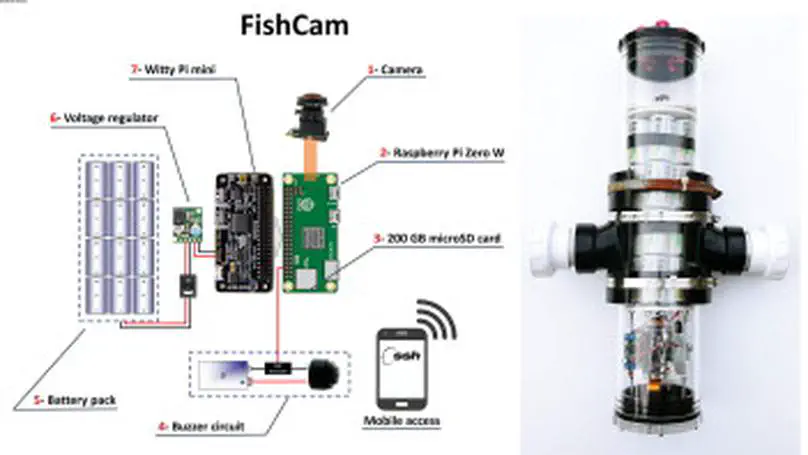
We describe the “FishCam”, a low-cost (500 USD) autonomous camera package to record videos and images underwater. The system is composed of easily accessible components and can be programmed to turn ON and OFF on customizable schedules.

neuTube is an open source software for reconstructing neurons from fluorescence microscope images. It is easy to use and improves the efficiency of reconstructing neuron structures accurately. The framework combines 2D/3D visualization, semi-automated tracing algorithms, and flexible editing options that simplify the task of neuron reconstruction.

Today, biologists spend too much time pipetting by hand. We think biologists should have robots to do pipetting for them. People doing science should be free of tedious benchwork and repetitive stress injuries.
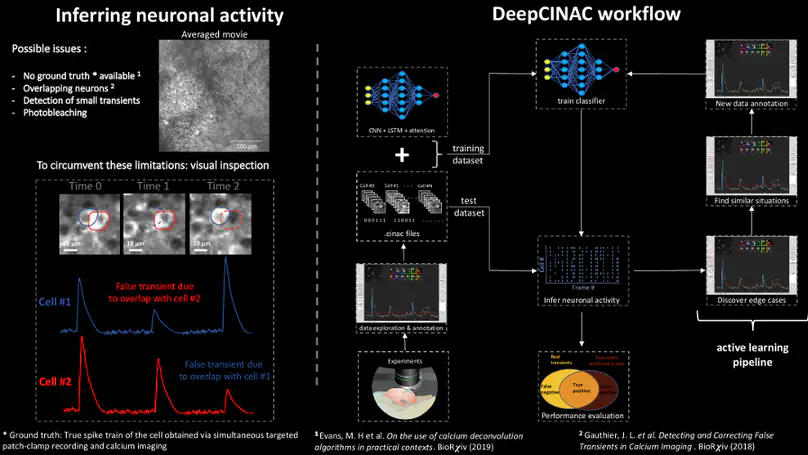
Two-photon calcium imaging is now widely used to infer neuronal dynamics from changes in fluorescence of an indicator. However, state of the art computational tools are not optimized for the reliable detection of fluorescence transients from highly synchronous neurons located in densely packed regions such as the CA1 pyramidal layer of the hippocampus during early postnatal stages of development.

The fingertip laser project makes use of the sensor used in an Avago ADNS-9500 laser mouse, to improve the capabilities of robotic hands, giving them the capability to detect distance, surface type and slippage of grasped objects.

SciDraw is a free repository of high quality drawings of animals, scientific setups, and anything that might be useful for scientific presentations and posters. We want this repository to be as open as possible, so do not require signing up to post a drawing.

The open-source optical toolbox UC2 [YouSeeToo] simplifies the process of building optical setups, by combining 3D-printed cubes, each holding a specific component (e.g. lens, mirror) on a magnetic square-grid baseplate. The use of widely available consumables and 3D printing, together with documentation and software, offers an extremely low-cost and accessible alternative for both education and research areas.