Posts
The US BRAIN Initiative archive for publishing and sharing neurophysiology data including electrophysiology, optophysiology, and behavioral time-series, and images from immunostaining experiments. DANDI: Distributed Archives for Neurophysiology Data Integration is a Web platform for scientists to share, collaborate, and process data from cellular neurophysiology experiments.
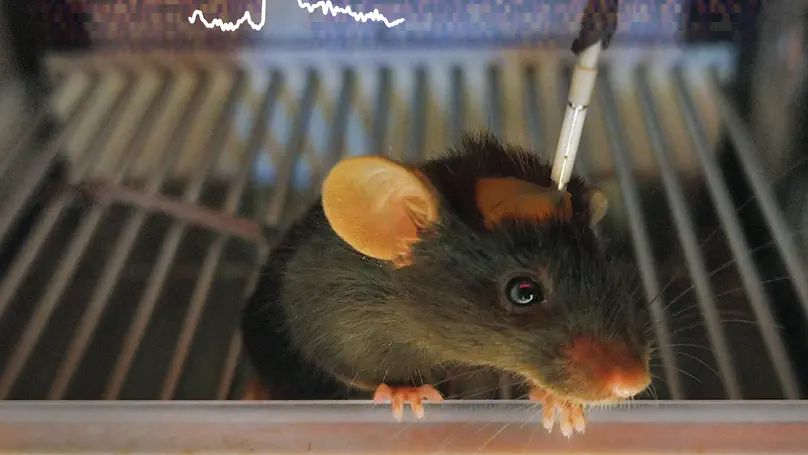
Fiber photometry (FP) is an adaptable method for recording in vivo neural activity in freely behaving animals. It has become a popular tool in neuroscience due to its ease of use, low cost, the ability to combine FP with freely moving behavior, among other advantages.
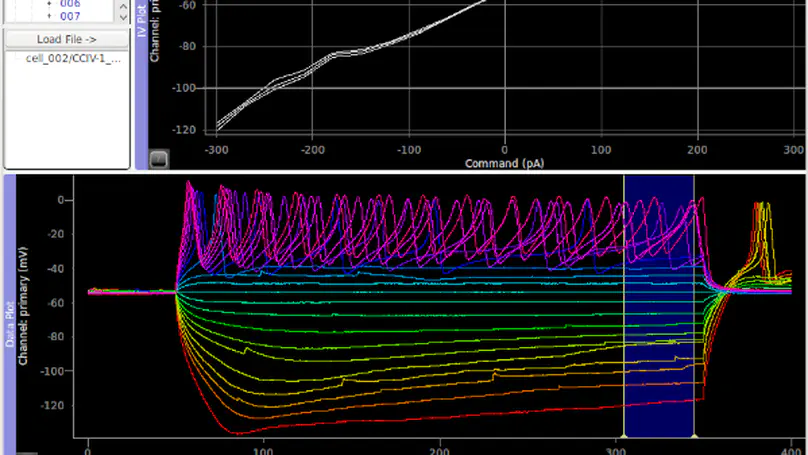
ACQ4 is a python-based platform for experimental neurophysiology developed at the Allen Institute for Brain Science and the University of North Carolina, Chapel Hill. It includes support for patch-clamp electrophysiology, multiphoton imaging, scanning laser photostimulation, and many other experimental techniques.
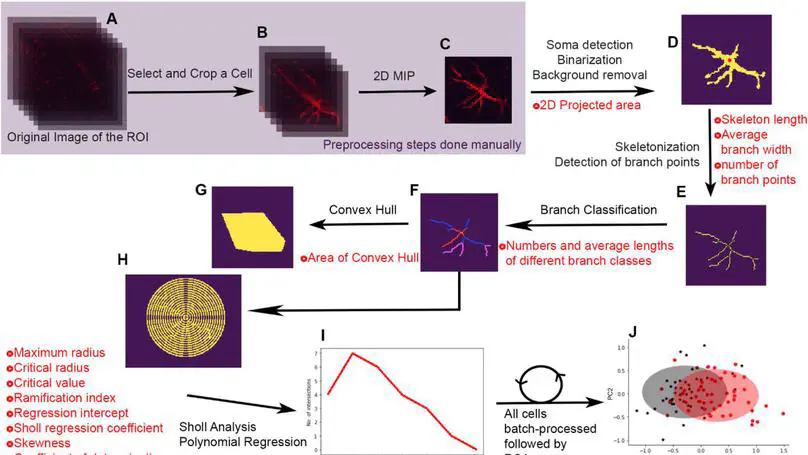
Nervous system development and plasticity involves changes in cellular morphology, making morphological analysis a valuable exercise in the study of nervous system development, function and disease. Morphological analysis is a time-consuming exercise requiring meticulous manual tracing of cellular contours and extensions.
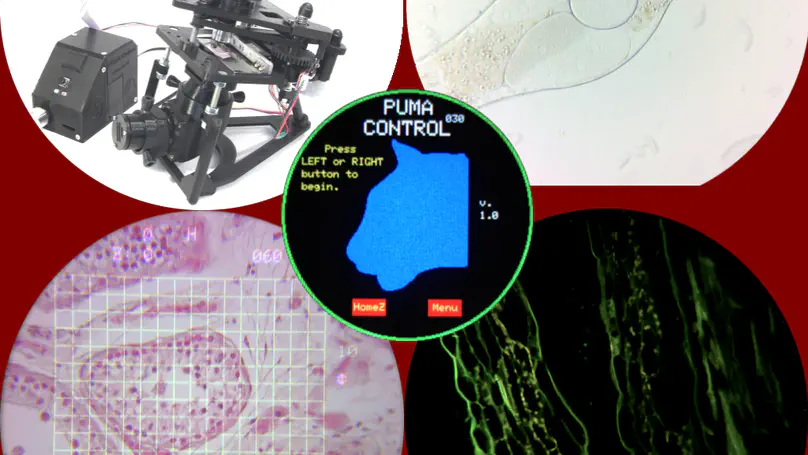
PUMA is 3D printed open source portable modular microscope with augmented reality, fluorescence, phase contrast, epi-illumination, epi- and trans-polarisation and other features. You can either build it fully DIY or purchase the basic scope ready made and upgrade it as required (see below for details).

One of the biggest challenges in neuroscience is to understand how neural circuits in the brain process, encode, store, and retrieve information. Meeting this challenge requires tools capable of recording and manipulating the activity of intact neural networks in freely behaving animals.
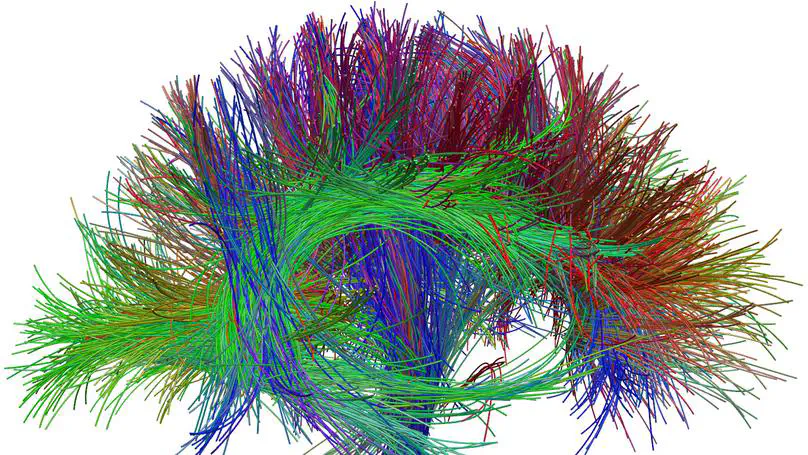
This is a diffusion-weighted MRI processing Matlab toolbox (including binaries), which can be used to: • Compute the Q-Ball Imaging Orientation Distribution Function in Constant Solid Angle (CSA-ODF) (Aganj et al, MRM 2010).
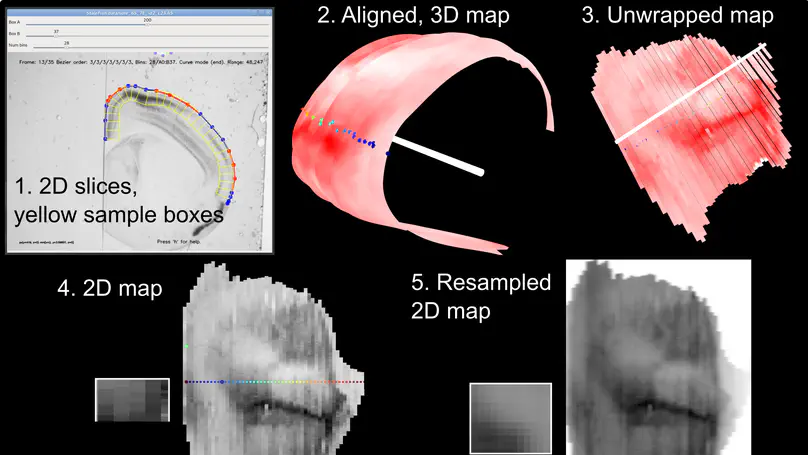
Stalefish is a tool to analyse gene expression from ISH or other histology slice images. From your 2D brain slice images you can build a 3D expression surface from a brain structure such as the isocortex or the hippocampus.

Autopilot is a Python framework for performing complex, hardware-intensive behavioral experiments with swarms of networked Raspberry Pis. As a tool, it provides researchers with a toolkit of flexible modules to design experiments without rigid programming & API limitations.
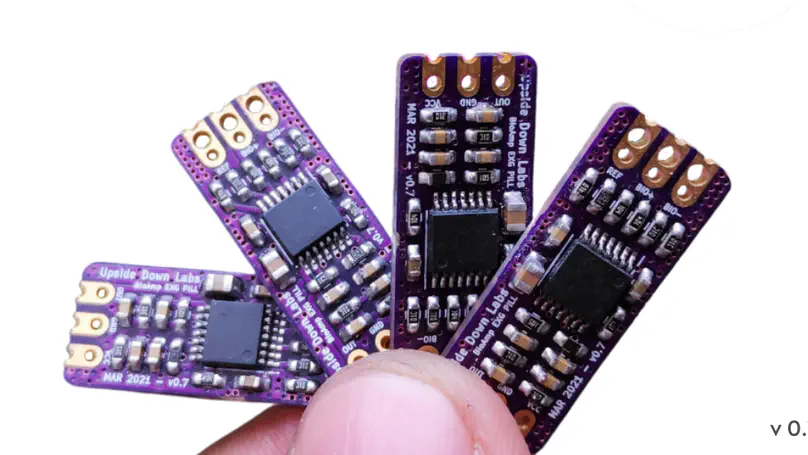
BioAmp EXG Pill is a small, powerful Analog Front End (AFE) biopotential signal acquisition board that can be paired with any 5 V Micro Controller Unit (MCU) with an ADC. It is capable of recording publication-quality biopotential signals like ECG, EMG, EOG, and EEG, without the inclusion of any dedicated hardware or software filters.