EASI-FISH
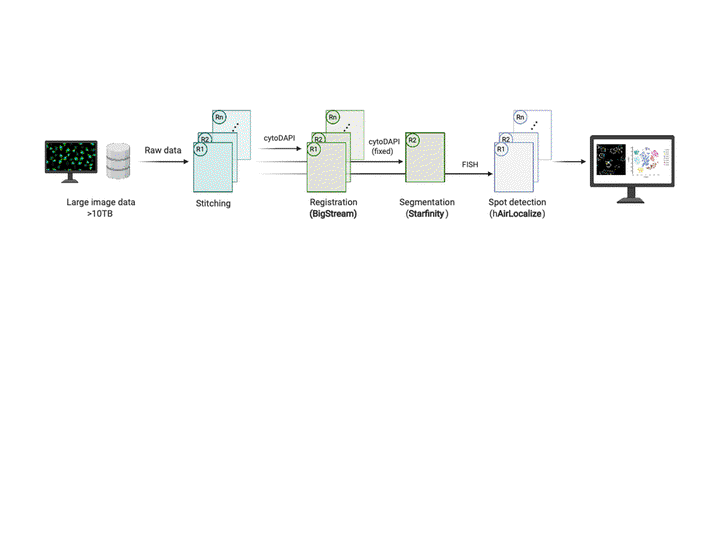
This workflow is used to analyze large-scale, multi-round, high-resolution image data acquired using EASI-FISH (Expansion-Assisted Iterative Fluorescence In Situ Hybridization). It takes advantage of the n5 filesystem to allow for rapid and parallel data reading and writing. It performs automated image stitching, distributed and highly accurate multi-round image registration, 3D cell segmentation, and distributed spot detection. We also envision this workflow being adapted for analysis of other image-based spatial transcriptomic data.
Check the pre-print here https://www.biorxiv.org/content/10.1101/2021.03.08.434304v1
Project Author(s)
Yuhan Wang; Konrad Rokicki; Avatar Cristian Goina
Project Links
https://github.com/multiFISH/EASI-FISH
This post was automatically generated by Matias Andina